xGen NGS—made to accelerate.
xGen Predesigned Hyb Panels are stocked enrichment panels for targeted next generation sequencing (NGS) research. The panels consist of individually synthesized and quality-controlled xGen Hyb Probes that are designed for use with the xGen Hybridization Capture Core Reagents to perform targeted sequencing.
Achieve uniform coverage and reliable capture results from a broad offering of xGen Predesigned Hyb Panels
Updated, automation-friendly protocol enables various levels of throughput
FIGYELEM!
IDT termékek szállítási költségei 2024-től:
A szintézis helyétől, valamint a szállítási körülményektől függően az alábbi költségek kerülnek felszámításra:
-
Belgiumból: nettó 6 900 Ft
USA-ból: nettó 34 000 Ft
Belgiumból, szárazjégen: nettó 37 000 Ft
USA-ból, szárazjégen: nettó 45 000 Ft
- Oldatok szállítási költsége: mennyiség és súly függvénye, kérjen ajánlatot a Bio-Science Kft.-től!
Általános tájékoztató az IDT USA gyártási központjából érkező „custom” termékcsoportokról:
szintetikus biológiai termékek:
- Gének
- gBlocks, eBlocks
- Ultramerek
- RNS-oligók
- CRISPR guide RNS-ek
- oPools
qPCR próbák:
a szintézis helye a szekvencia komplexitásától, valamint a választott festék/ quencher kombinációtól függ
szolgáltatások:
- PAGE tisztított termékek
stock termékek:
nem áll teljes lista rendelkezésre, kérjen ajánlatot a Bio-Science Kft.-től!
xGen™ Exome Hybridization Panel
Higher on-target percentage—even with a 12-plex capture
The xGen Exome Hyb Panel v2 is designed using a target-aware algorithm that reduces off-target binding while maximizing coverage. We manufacture this panel under ISO 13485 standards as a single large lot, promoting equal representation and consistency from one aliquot to the next.
xGen NGS—made for whole exome sequencing.
xGen™ Acute Myeloid Leukemia (AML) Cancer Hybridization Panel
Predesigned panel that targets AML-associated genes
The xGen Acute Myeloid Leukemia (AML) Cancer Hyb Panel comprises 11,731 xGen Hyb Probes, spanning 1.19 Mb of the human genome, for research using targeted enrichment of >260 AML-associated genes, for more efficient identification of disease-causing mutations in research samples.
xGen NGS—made for acute myeloid leukemia research.
xGen™ Inherited Diseases Hybridization Panel
Targeted sequencing research on genes associated with inherited diseases
The xGen Inherited Diseases Hyb Panel enables research involving deeper sequencing of genomic regions containing genes and SNPs associated with inherited diseases. The gene list is based on the HGMD® repository of known inherited disease-causing mutations.
xGen NGS—made for inherited diseases research.
xGen™ Pan-Cancer Hybridization Panel
Research the genes implicated in several cancers
The xGen Pan-Cancer Hyb Panel consists of 7,816 xGen Hyb Probes for research, spanning 800 kb of the human genome, for enrichment of 127 mutated genes implicated across 12 tumor tissue types for deeper sequencing coverage in research studies.
xGen NGS—made for cancer research.
xGen™ Human Copy Number Variant (CNV) Backbone Hybridization Panel
Research copy number variations in the human genome
The xGen Human CNV Backbone Hyb Panel spans the entire genome, allowing you to generate reference coverage data that can be used for research on copy number variations (CNVs). Use this as a standalone panel or a spike-in to other xGen Hyb Panels.
xGen™ NGS—made for variant research.
xGen™ Human Identification (ID) Hybridization Panel
Uniquely identify research samples by SNPs
The IDT xGen™ Human ID Hyb Panel enables unique identification of individual samples from a larger population. The panel provides genomic target enrichment of a distinctive set of single nucleotide polymorphisms (SNPs), including high minor allele frequency SNPs and a set of 5 gender marker SNPs. With a power of discrimination greater than 1 in 5 million, the xGen Human ID Hyb Panel is compatible with various sample types, including cell-free DNA and DNA from formalin-fixed, paraffin-embedded (FFPE) tissues.
xGen NGS—made for human gene identification research.
xGen™ Human Mitochondrial DNA (mtDNA) Hybridization Panel
Complete coverage of the human mitochondrial genome
The xGen Human Mitochondrial DNA (mtDNA) Hyb Panel is designed to simplify the investigation of human mitochondrial genes by delivering optimal coverage across the entire >16 kb human mitochondrial genome in research samples.
xGen NGS—made for human mtDNA research.
xGen™ SARS-CoV-2 Hybridization Panel
Strain-agnostic enrichment of SARS-Cov-2 genome
The xGen SARS-CoV-2 Hyb Panel spans the complete viral genome and is not susceptible to known and novel mutations arising from diverse strains.
xGen NGS—made for SARS-CoV-2 research.
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend for these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.
Consistent data from one aliquot to the next
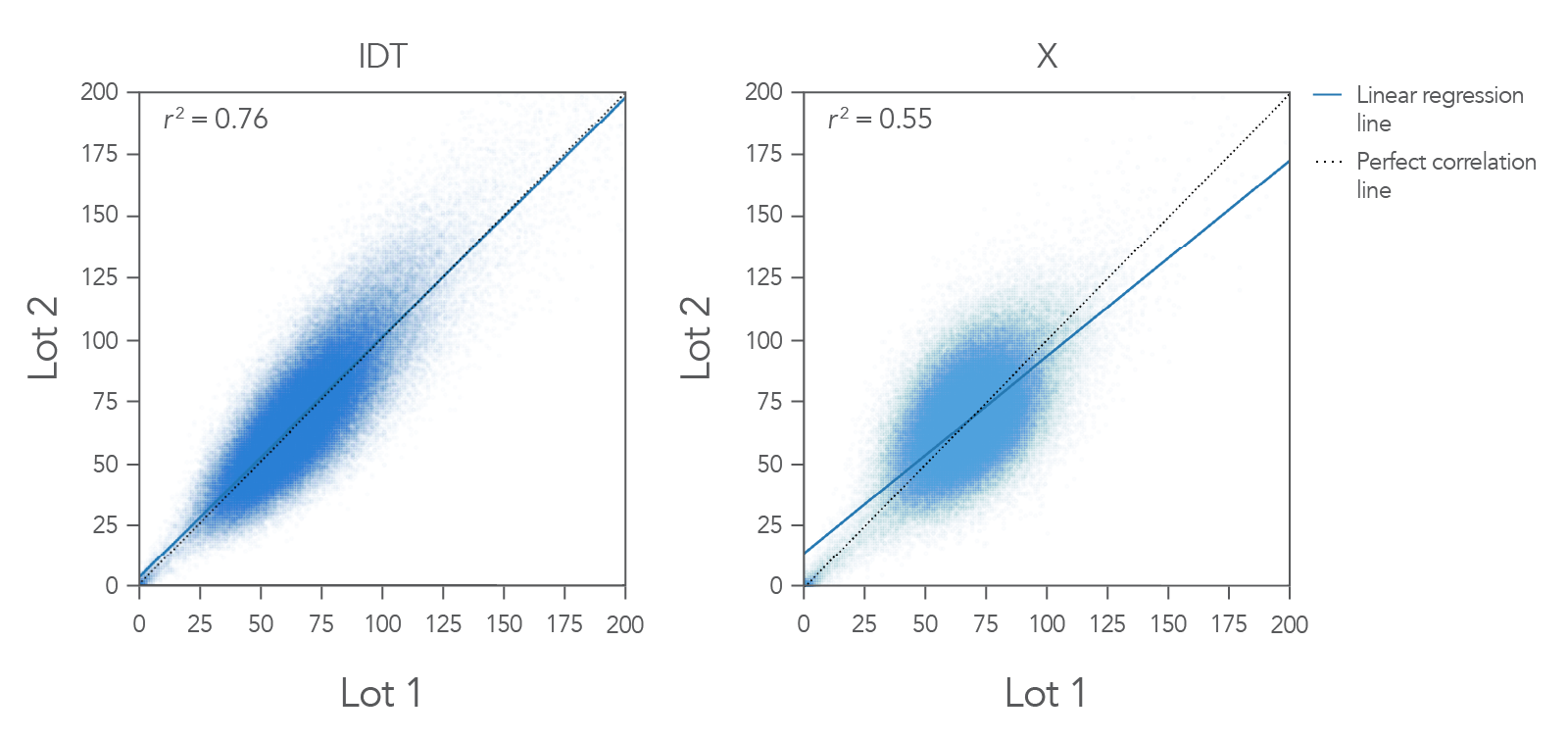
Figure 1. Example coverage of the IDT xGen Exome Hyb Panel v2 vs. competitor. Two different users performed hyb captures on different days in different locations using different aliquots (lots) of the IDT xGen Exome Hyb Panel v2. Captures were performed using 100 ng Coriell DNA NA12878 in a singleplex reaction (n=3). Bioinformatics analysis was done for 3-way coverage correlation comparison, a representative pair from each vendor is demonstrated here. The IDT xGen Exome Hyb Panel v2 shows a linear regression line that mimics the predicted one-to-one correlation line with an r2 value of 0.76. In this example, IDT outperformed the competitor.
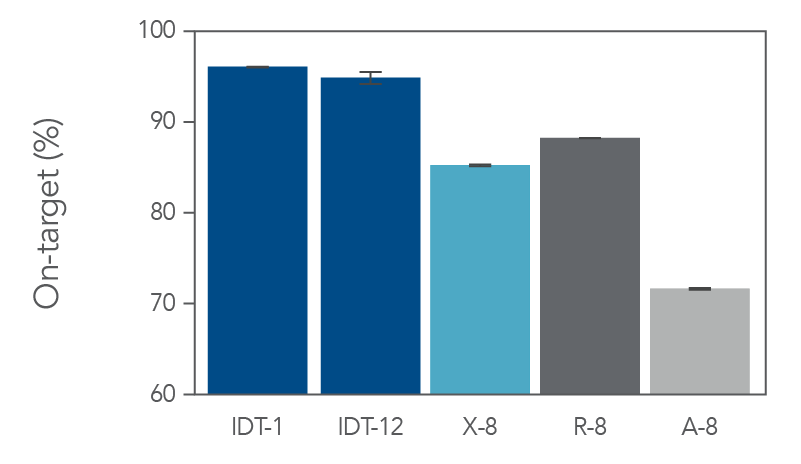
On-target coverage and uniformity of the xGen Exome Hyb Panel v2
Figure 2. The IDT whole exome sequencing workflow has higher on-target percentage in comparison to alternate suppliers. Sequencing libraries were prepared using enzymatic shearing and ligation-based library preparation using the library prep protocols from each vendor. Libraries were captured and multiplexed according to the vendor’s exome panel capture protocol (X-8, R-8, and A-8, where “8” signifies 8-plex captures, n=2). For IDT, we show both 1-plex (n=3) and 12-plex (n=2) captures (IDT-1 and IDT-12, respectively). Enriched libraries were sequenced on a NextSeq® instrument (Illumina) in high output mode using 2 x 100 bp paired-end reads for analysis. On-target bases were determined with Picard (percent selected bases) using 5 Gb per library.
Figure 3. Consistent uniform sequence coverage with the xGen Exome Hyb Panel v2. DNA libraries were created from 100 ng of human genomic DNA (Coriell) and enriched either as 1-plex captures (n=3) or as a single 12-plex (n=2) capture using the xGen Exome Hyb Panel v2. The enriched libraries were sequenced (2 x 100 bp reads) on a NextSeq® instrument (Illumina) and subsampled to 5 Gb. The data shows uniform coverage with a flanked on-target rate of 94.7%, mean target coverage of 64.5X, and a duplication rate of 3.3% (calculated with Picard).
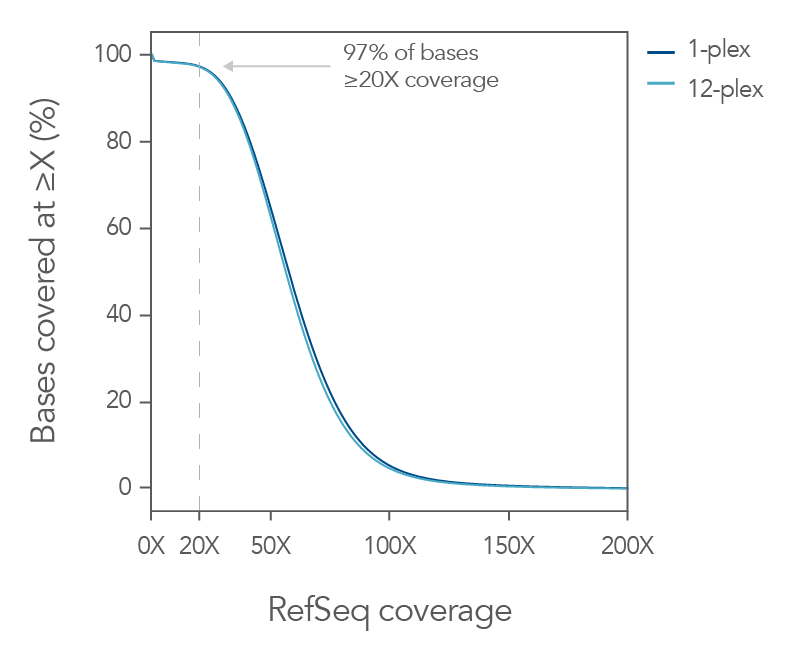
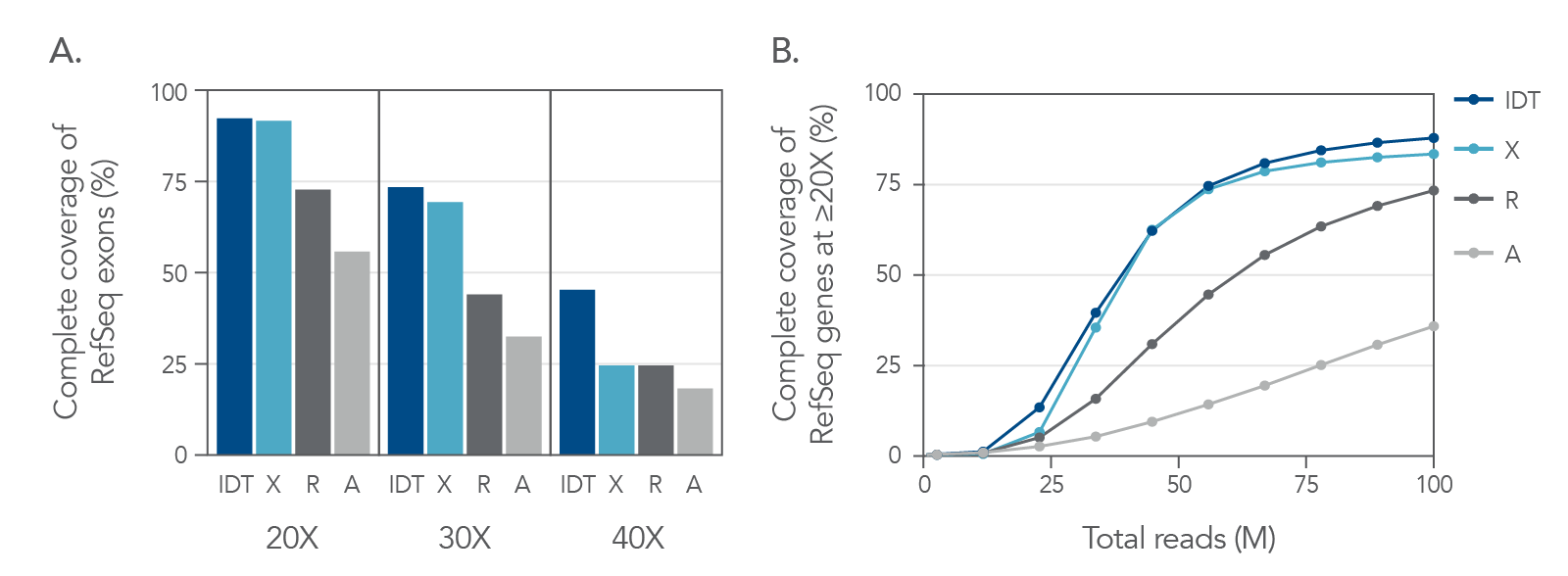
Coverage of RefSeq database with minimal sequencing
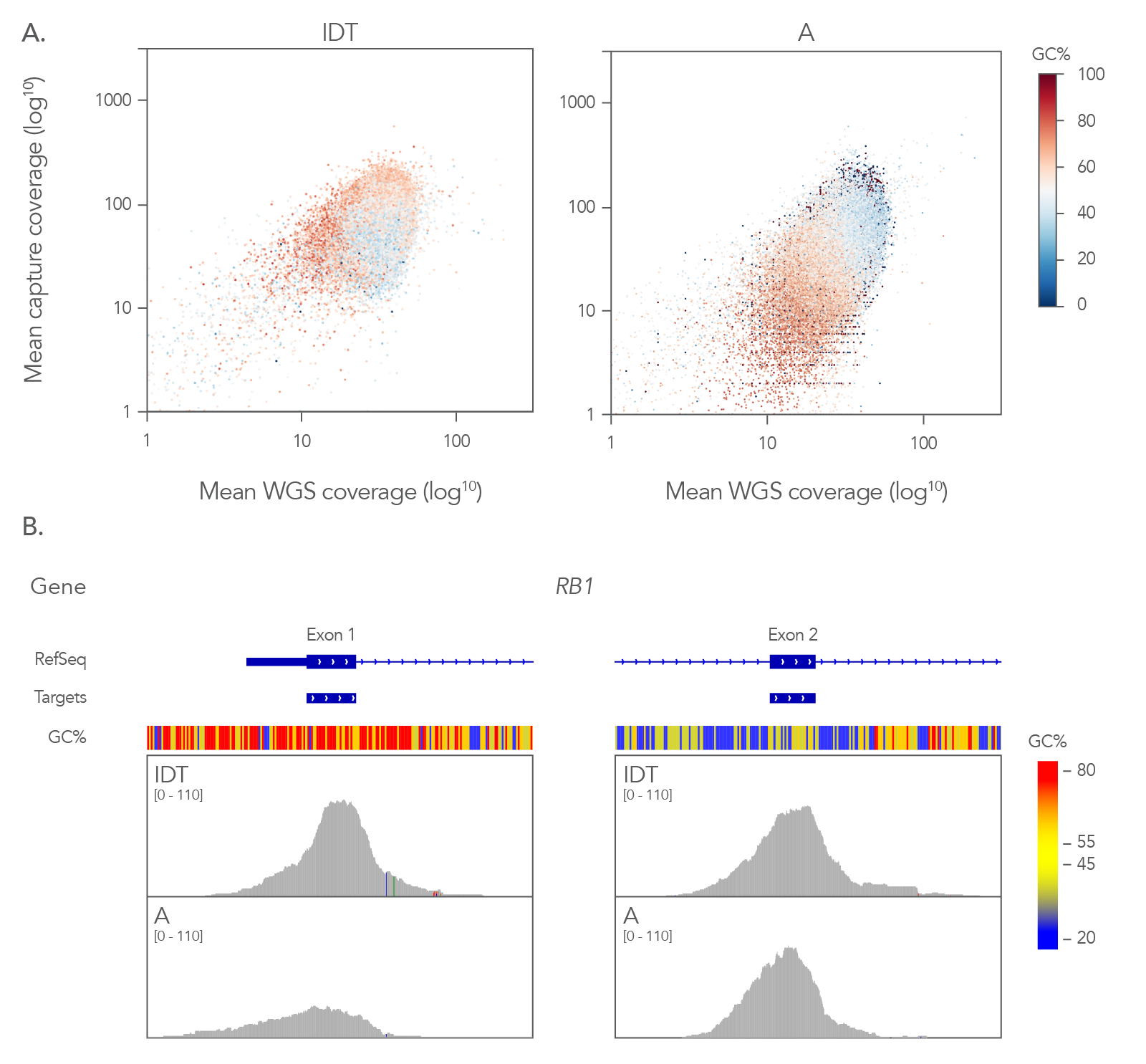
Figure 4. Coverage profile of the xGen Exome Hyb Panel v2 closely resembles whole genome data. (A) Comparing the coverage depths of different exome panels (8-plex, n=1) to coverage depth from whole genome sequencing (WGS) of a matched library shows that the xGen Exome Hyb Panel v2 closely matches WGS. Analysis of guanine-cytosine content (GC) shows that the xGen Exome Hyb Panel has a more effective capture of GC content (compare dark red and red dots in the right and left panels). (B) RB1 exons 1 and 2 show extremes of GC content with ~76% in exon 1 and ~38% in exon 2. A comparison of capture between IDT and supplier A shows a higher read depth across exon 1 for the xGen Exome Hyb Panel v2 on the Integrative Genomics Viewer (Broad Institute), whereas supplier A coverage has a lower number of reads of this exon with high GC content.
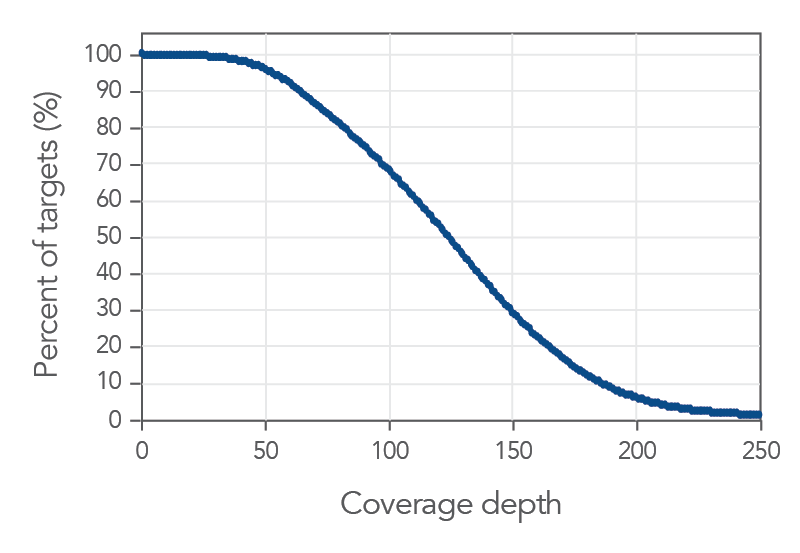
Figure 5. Exome coverage of the xGen Exome Hyb Panel v2. (A) 100 ng Coriell gDNA hybridization capture libraries (n=2) were sequenced with 5 Gb per sample while the percent of exons covered end-to-end at each read depth were calculated. The xGen Exome Hyb Panel v2 (IDT) shows the highest percentage of exons covered at each indicated depth (compared to suppliers X, R, and A). (B) The xGen Exome Hyb Panel v2 provides end-to-end RefSeq gene coverage at ≥20X. Individual samples were subsampled at different read depths (2 x 100 bp read length). The percentage of genes that were covered for every base of every exon at ≥20X was calculated at each read depth and plotted.
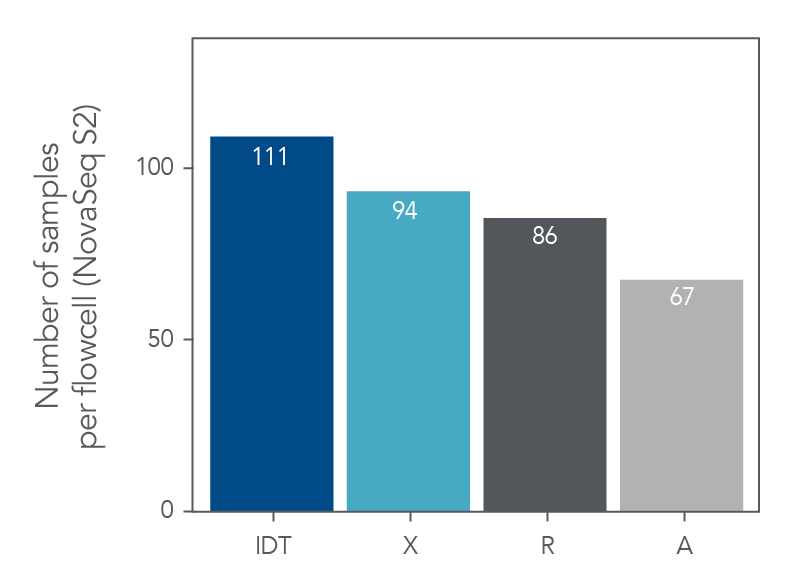
Maximize the number of samples per flowcell
Figure 6. xGen Exome Hyb Panel v2 can increase the number of samples loaded into a flowcell. DNA libraries were created from 100 ng of human genomic DNA (Coriell) and enriched either as 8-plex, n=1 (supplier R), 8-plex, n=2 (suppliers X and A) or 12-plex, n=2 (IDT) captures. The enriched libraries were sequenced (2 x 100 bp) on a NextSeq® instrument (Illumina) and the number of reads required to achieve 75X mean target coverage (Picard) per sample was estimated along with the number of samples that would fit on a NovaSeq™ S2 flowcell (Illumina).
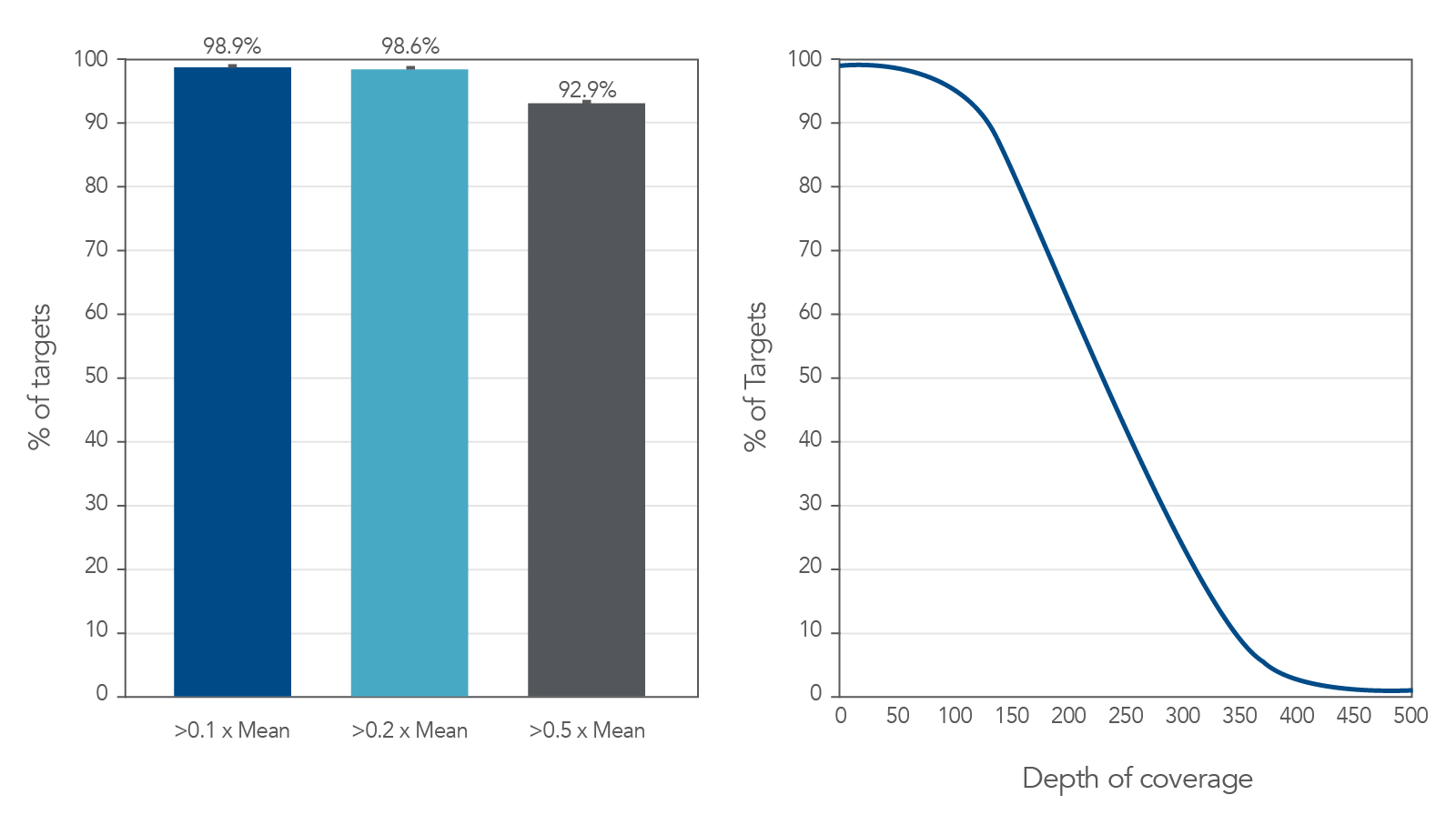
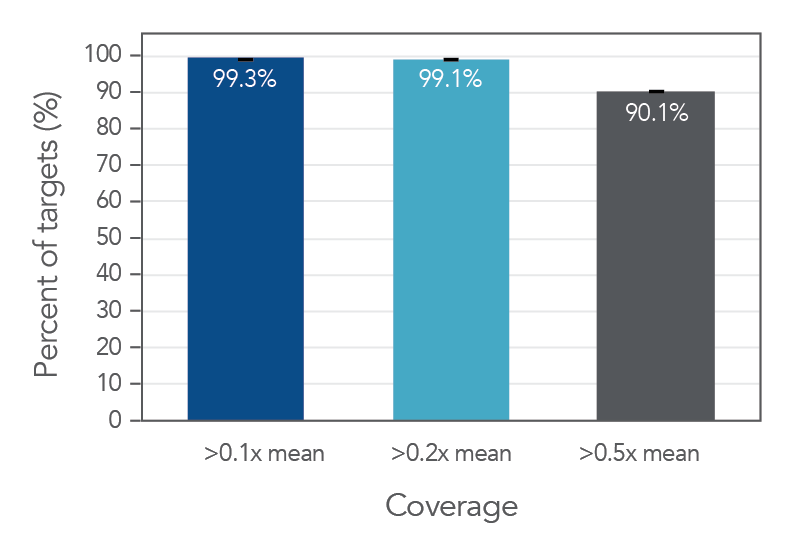
Coverage uniformity for the xGen AML Cancer Hyb Panel
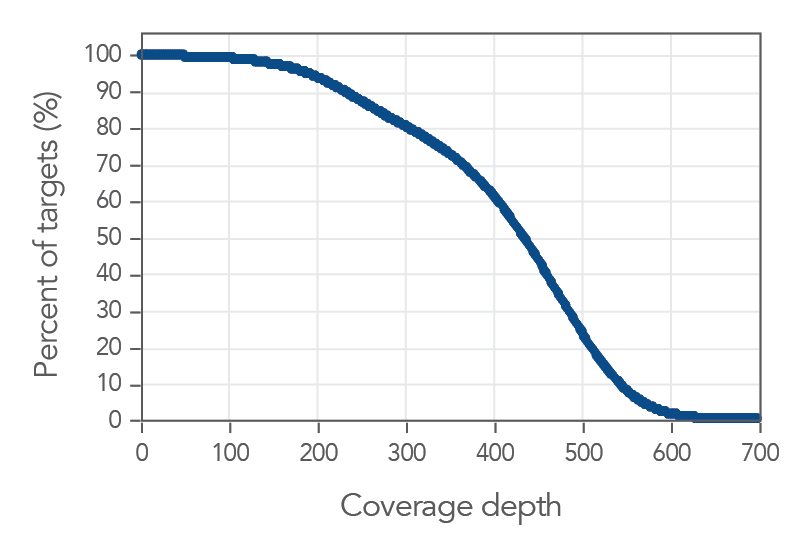
Figure 1. Coverage uniformity obtained with xGen AML Cancer Panel. Greater than 0.2X mean coverage is observed for >98% of targets. Illumina TruSeq® HT libraries (n=8) were enriched using the xGen AML Cancer Panel and sequenced on a NextSeq 550 system using 2 x 150 paired-end reads. Each sample was sub-sampled to 4M total reads.
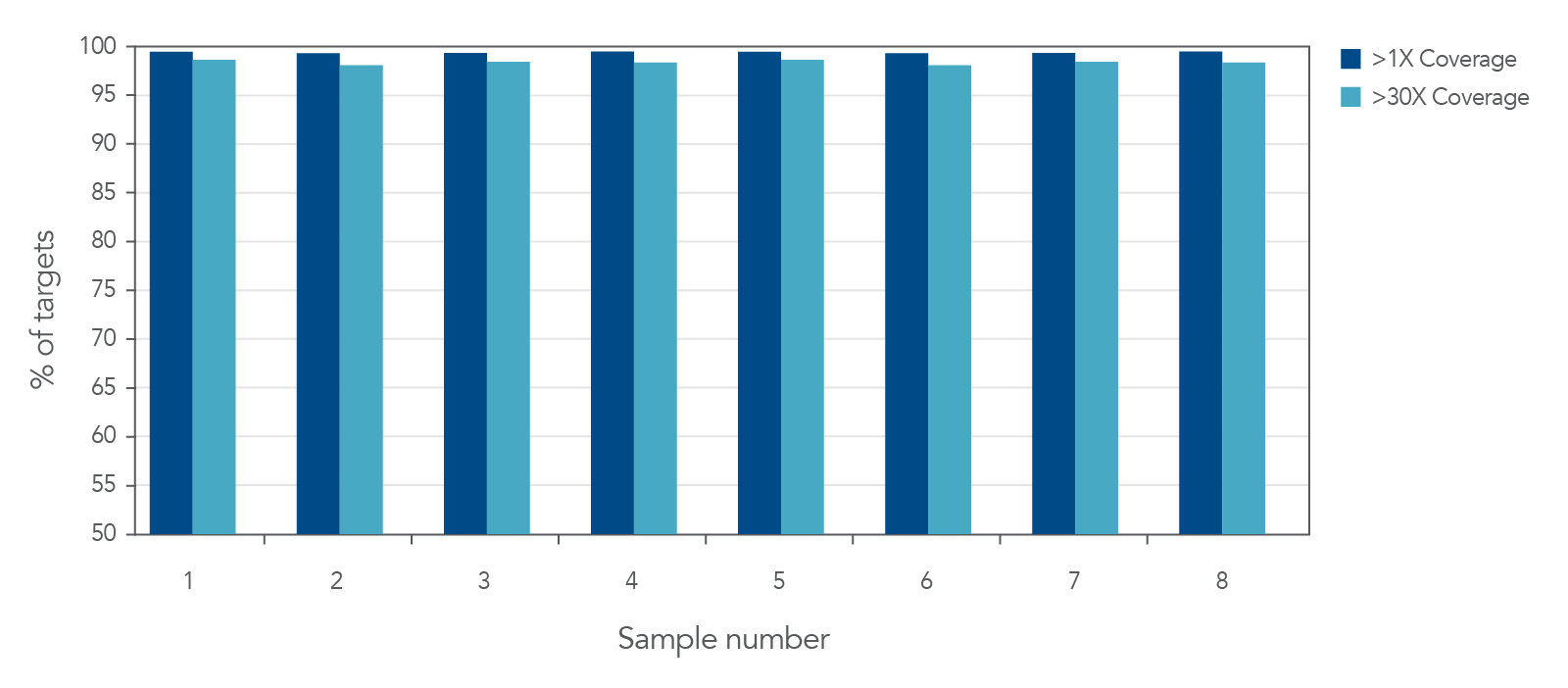
Figure 2. Deep coverage of targeted regions using xGen AML Cancer Panel. Eight genomic DNA libraries were enriched using the xGen Acute Myeloid Leukemia Cancer Panel and sequenced on a NextSeq 550 system using 2 x 150 paired-end reads. Total reads for each sample were sub-sampled to 4M total reads. In all samples, there was >1X coverage for 99.3% of targets and >30X coverage for 98.7% of targets.
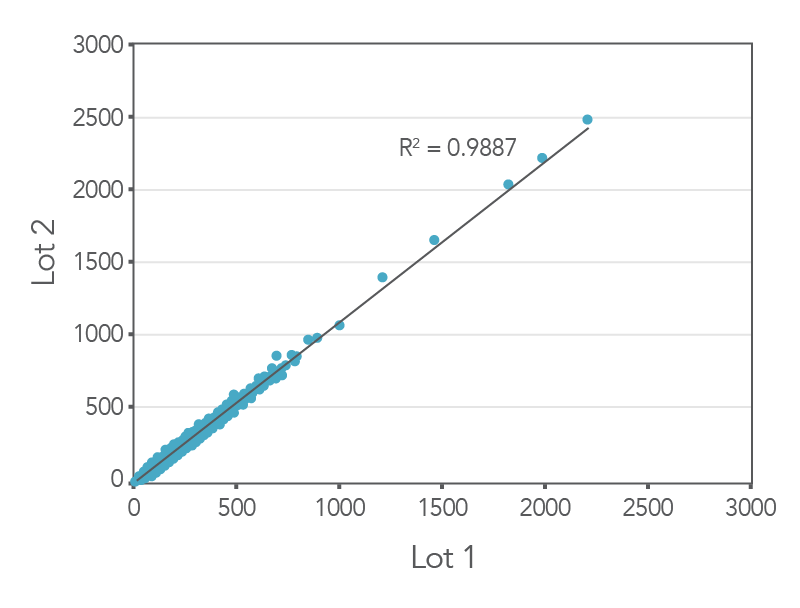
Consistent results between lots
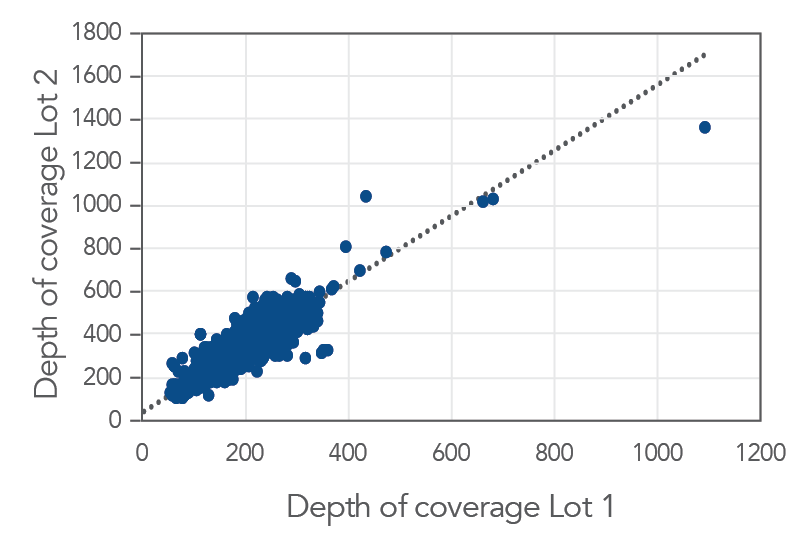
Figure 3. Consistent results obtained with xGen AML Cancer Panel. Genomic DNA libraries were enriched with two lots of the xGen AML Cancer Panel (n=8 for each lot). The enriched samples were sequenced on the NextSeq 550 sequencing platform using 2 x150 paired-end reads. All samples were sub-sampled to a level of 4M total reads. The probe-by-probe target coverage was averaged for eight replicate samples for each lot of the xGen AML Cancer panel. A comparison of probe-by-probe target coverage between two lots show consistent results between lots, with an R2 value of 0.9887.
Uniformity of coverage for xGen Inherited Diseases Hyb Panel
Figure 1. An example of high coverage uniformity obtained with xGen Inherited Diseases Hyb Panel. Greater than 0.2X mean coverage is observed for >99% of targets. Illumina TruSeq® HT libraries (n=8) were enriched using the xGen Inherited Diseases Hyb Panel and sequenced on a NextSeq® 550 system using 2 x 150 paired-end reads. Each sample was sub-sampled to 17.5M total reads.
Complete coverage across all targets
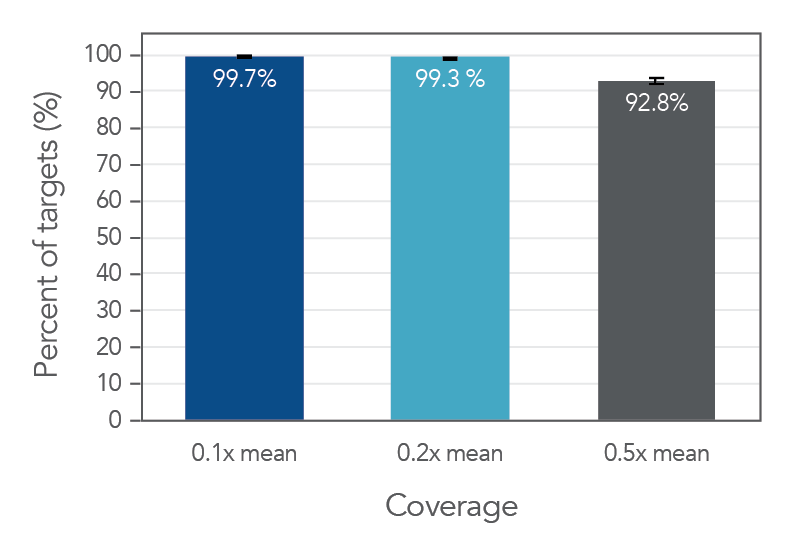
Figure 2. An example of deep coverage of targeted regions using xGen Inherited Diseases Hyb Panel. Eight genomic DNA libraries were enriched using the xGen Inherited Diseases Hyb Panel and sequenced on a NextSeq 550 system using 2 x 150 paired-end reads. Total reads for each sample were sub-sampled to 17.5M total reads. In all samples, there was >1X coverage for 99.4% of targets and >30X coverage for 98.8% of targets.
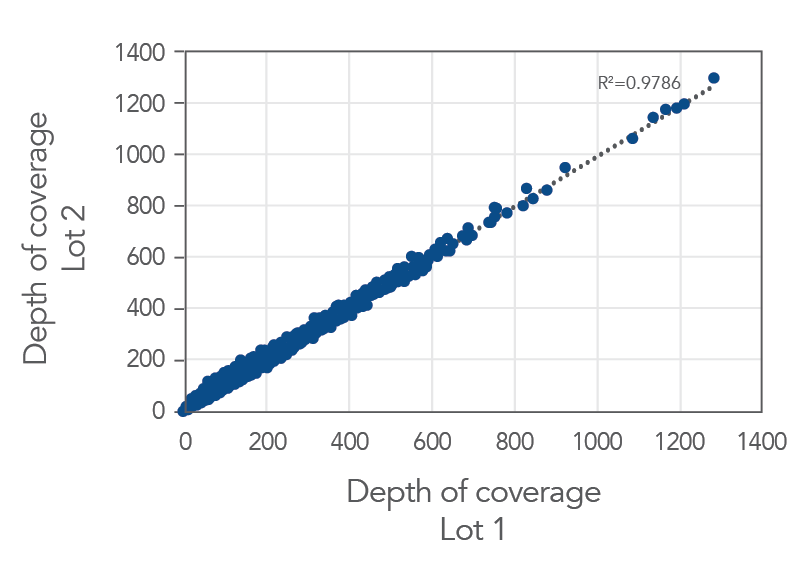
Consistent data from one lot to the next
Figure 3. An example of lot-to-lot consistency obtained with xGen Inherited Diseases Hyb Panel. Genomic DNA libraries were enriched with two lots of the xGen Inherited Diseases Hyb Panel (n=8 for each lot). The enriched samples were sequenced on the NextSeq 550 sequencing platform using 2 x150 paired-end reads. All samples were sub-sampled to a level of 17.5M total reads. The probe-by-probe target coverage was averaged for eight replicate samples for each lot of the xGen Inherited Diseases Hyb Panel. A comparison of probe-by-probe target coverage between two lots showed excellent reproducibility, with an R2 value of 0.9786.
30X coverage depth for 99% of the targets
Figure 1. An example of, deep coverage of targeted regions using xGen Pan-Cancer Hyb Panel. Genomic DNA libraries (n=8) were enriched using the xGen Pan-Cancer Hyb Panel and sequenced on a NextSeq® 550 system using 2 x 150 paired-end reads. Total reads for each sample were sub-sampled to 4M total reads. In all samples, there was >1X coverage for 99.9% of targets and >30X coverage for 99.7% of targets.
Coverage uniformity for the xGen Pan-Cancer Hyb Panel
Figure 2. An example of high coverage uniformity obtained with xGen Pan-Cancer Hyb Panel. Greater than 0.2X mean coverage is observed for >99.3% of targets. Genomic DNA Illumina TruSeq® HT libraries (n=8) were enriched using the xGen Pan-Cancer Hyb Panel and sequenced on a NextSeq® 550 system using 2 x 150 paired-end reads. Each sample was sub-sampled to 4M total reads.
Consistent data from one lot to the next
Figure 3. An example of lot to lot consistency obtained with xGen Pan-Cancer Hyb Panel. Genomic DNA libraries were enriched with two lots of the xGen Pan-Cancer Hyb Panel (n=8 for each lot). The enriched samples were sequenced on the NextSeq 550 sequencing platform using 2 x150 paired-end reads. All samples were sub-sampled to a level of 4M total reads. The probe-by-probe target coverage was averaged for eight replicate samples for each lot of the xGen Pan-Cancer Hyb Panel. A comparison of probe-by-probe target coverage between two lots showed excellent consistency, with an R2value of 0.8378.
xGen Human ID Hyb Panel as a spike-in for sample identification
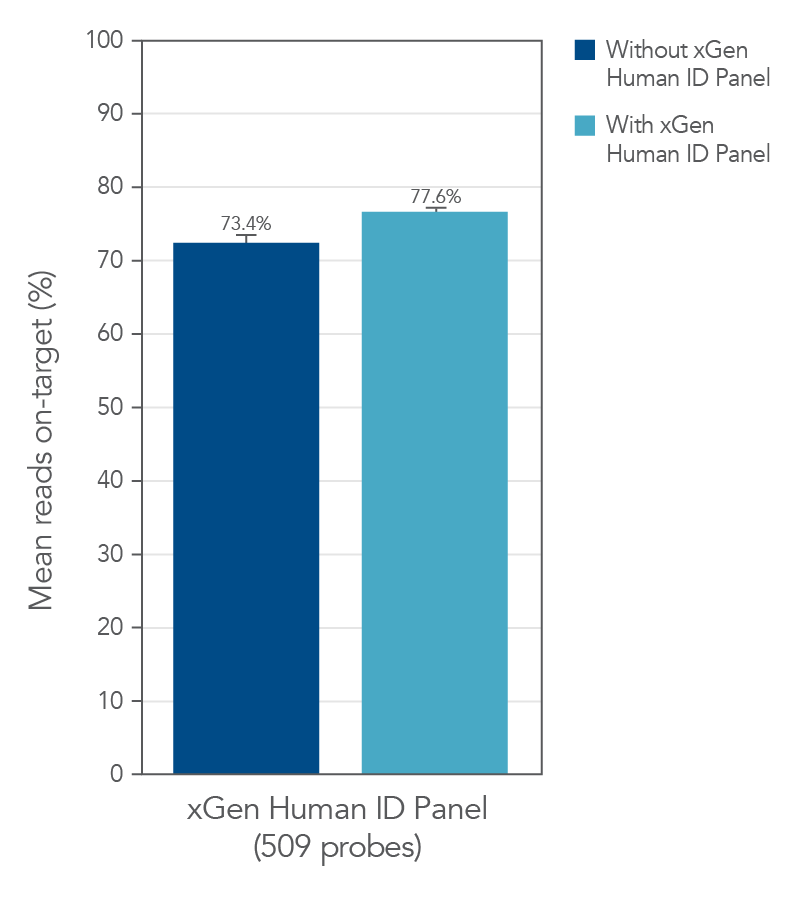
The xGen Human ID Hyb Panel can be used as a spike-in panel to larger capture panels in research studies. The xGen Human ID Hyb Panel does not affect target specificity and coverage uniformity of a representative xGen Custom Hyb Panel (Figure 1 and Figure 2).
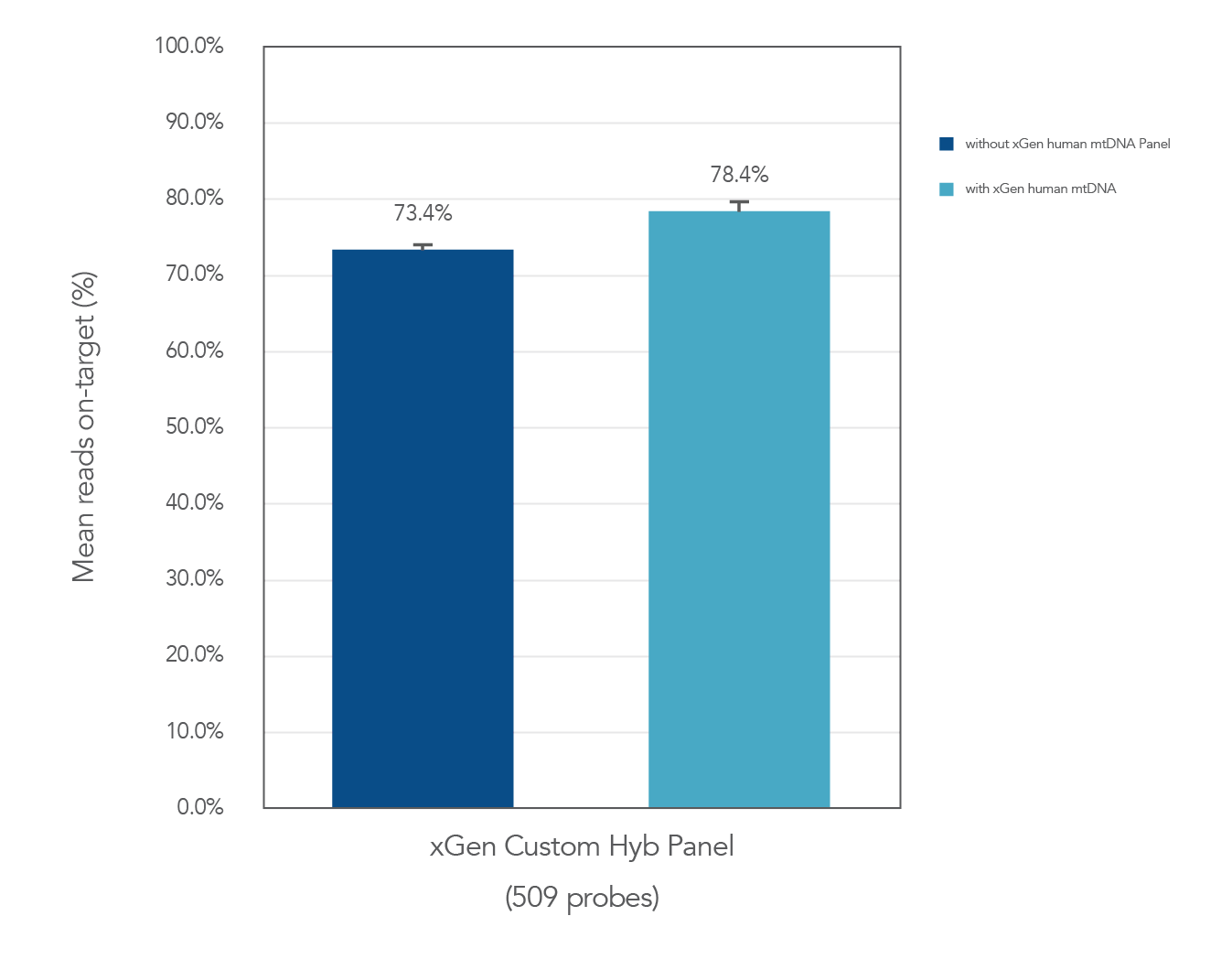
Figure 1. Example showing the spike in of the xGen Human ID Hyb Panel does not impact on-target performance of recipient panel. DNA libraries were prepared from NA12878 genomic DNA. 500 ng of the libraries were enriched with a custom panel comprised of 509 xGen Hyb Probes, with (n=8) and without spike in (n=3) of the xGen Human ID Research Panel. The enriched libraries were sequenced on a MiSeq® System (Illumina) using 2 X 150 paired-end reads. Each sample was sub-sampled to 0.5M total reads. The average of on-target rate (with 150 bp flank) across experiments are shown.
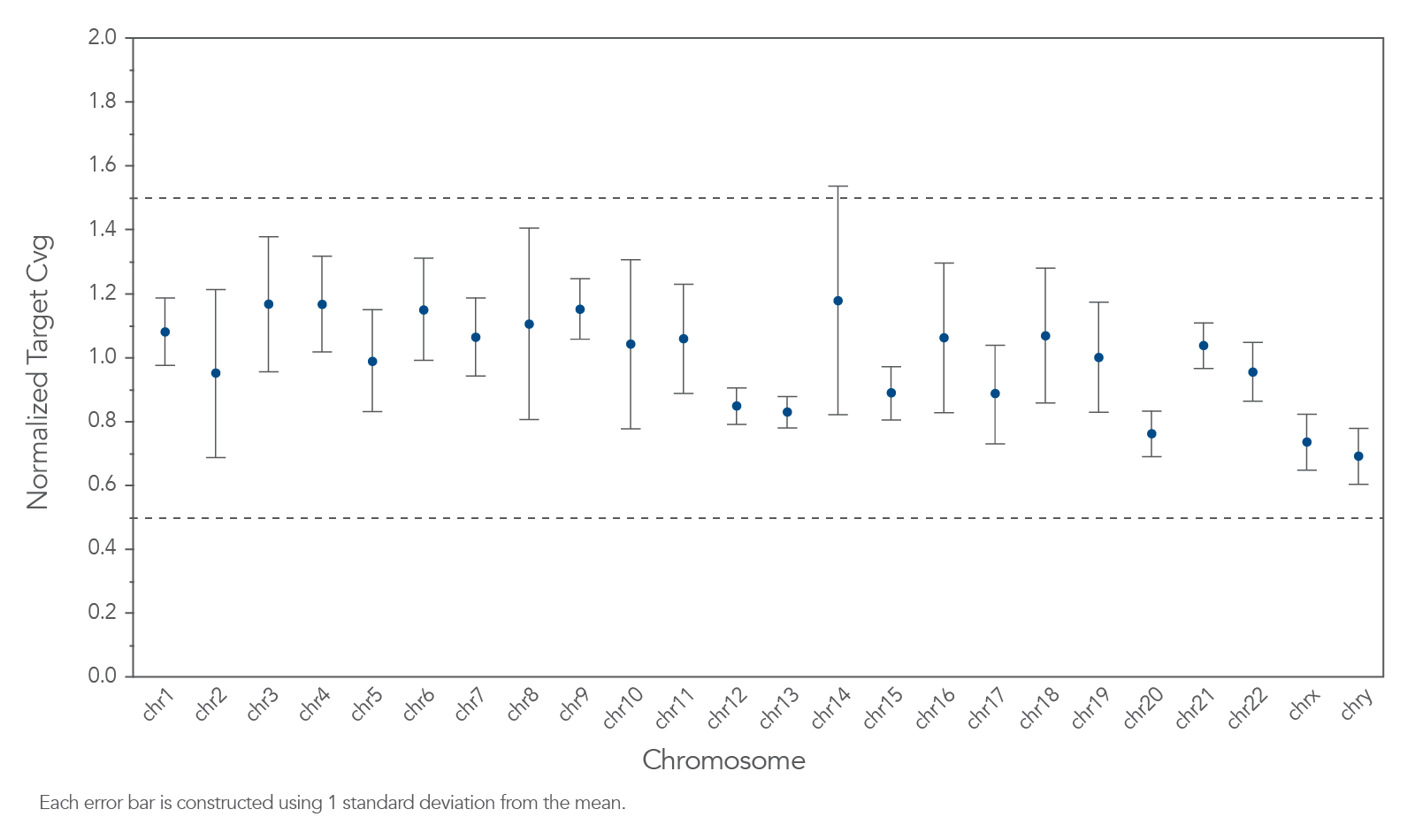
Figure 2. Example of uniform coverage of targeted regions using xGen Human ID Hyb Panel. Genomic DNA libraries were enriched using the xGen Human ID Hyb Panel spiked into a custom panel containing 509 probes (n=8). Enriched samples were sequenced on a MiSeq system using 2 x 150 paired-end reads. Total reads for each sample were sub-sampled to 0.5M total reads. The average normalized target coverage ranged between 0.60X and 1.51X for the regions targeted by the human ID Panel. Normalized target coverage bounds at 0.5X and 1.5X are denoted by the doted lines.
Efficient capture with combined panels
Figure 1. An example showing the spike in of the xGen Human mtDNA Hyb Panel does not impact on-target rate of recipient panel. DNA libraries were prepared from 100 ng of NA12878 genomic DNA (Coriell). 500 ng of the libraries were enriched with a custom panel comprised of 509 xGen Hyb Probes, with (n=8) and without spike in (n=3) of the xGen mtDNA Hyb Panel. The enriched libraries were sequenced on a MiSeq® System (Illumina) using 2 x 150 paired-end reads. Each sample was sub-sampled to 0.5M total reads. The averages of on-target values (with 150 bp flank) across experiments are shown.
Uniform coverage of targeted regions
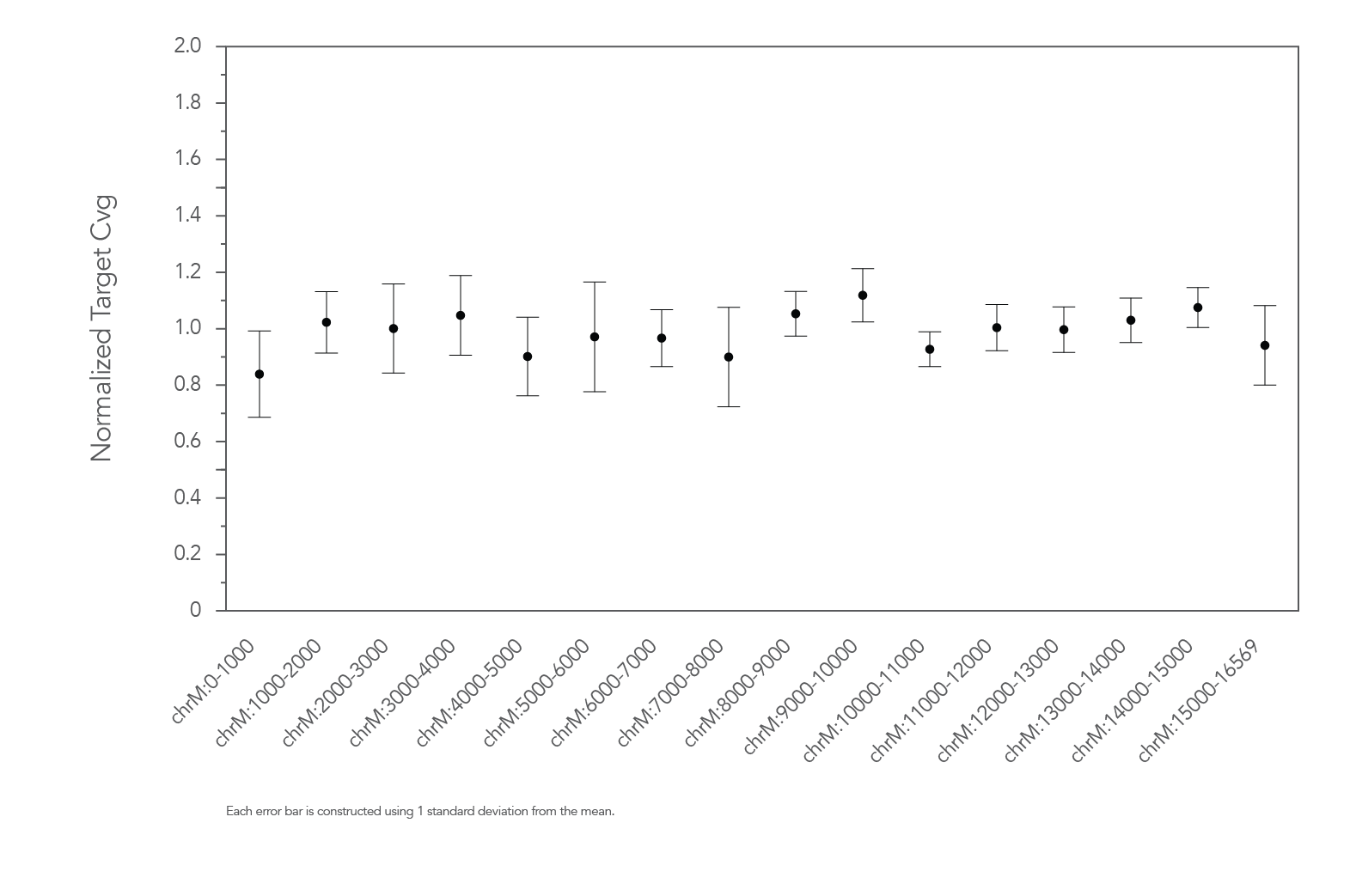
Figure 2. Example of uniform coverage of targeted regions using xGen Human mtDNA Hyb Panel. Genomic DNA libraries were enriched using the human mtDNA Panel spiked into a custom panel containing 509 probes (n=8). Enriched samples were sequenced on a MiSeq system using 2 x 150 paired-end reads. Total reads for each sample were sub-sampled to 0.5M total reads. The average normalized target coverage ranged between 0.56X and 1.31X for the regions targeted by the mtDNA Panel. Normalized target coverage bounds at 0.5X and 1.5X are denoted by the doted lines.
Even coverage across the complete viral genome
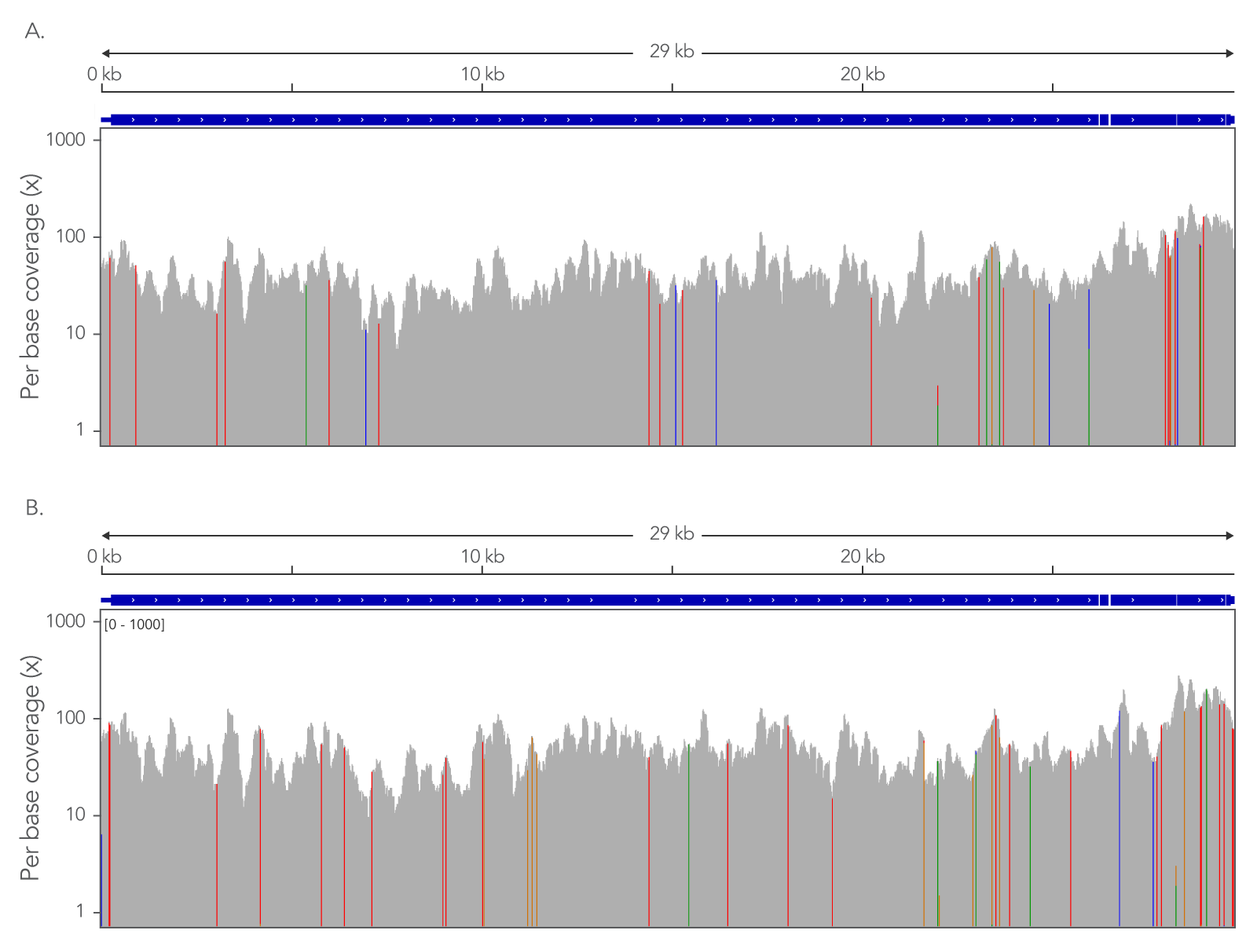
Figure 1. An example of the xGen™ SARS-CoV-2 Hyb Panel providing even coverage across the complete viral genome allowing strain identification with minimal read depth. (A) Coverage of a nasopharyngeal swap sample identified as belonging to strain Alpha (B.1.1.7). (B) Coverage of a nasopharyngeal swap sample identified as belonging to strain Delta (B.1.617.2). 5 µL of total RNA sample (previously shown to contain viral sequence via qPCR assay) were used to prepare NGS libraries (n=7) using the xGen™ Broad Range Library Prep Kit. Libraries were quantified and equal amounts were captured with the xGen™ Hybridization and Wash Kit. Samples were sequenced on an Illumina MiSeq™ and subsampled to 25000 total reads per sample. Data was aligned to the Refseq sequence (NC_045512) and viewed in the Integrated Genome Viewer (IGV). The IGV view color codes bases that do not match the canonical sequence. Strains were identified by submitting a consensus sequence for the sample to https://pangolin.cog-uk.io/
Exome sequencing targets only the protein-coding regions of the genome. Since exons represent only 1% of the human genome, a method of separating these regions from noncoding DNA is important to study mutations that could be important for disease research. The captured genomic material must also be suitable for sequencing to a satisfactory depth of coverage for research on variant alleles and sensitive applications like copy number variation (CNV) detection. To provide this increased depth of coverage and enable high multiplexing of samples, the xGen Exome Hyb Panel v2 targets only the coding sequences (CDS) of human coding genes in the RefSeq 109 database.
Our xGen Exome Hyb Panel v2 consists of 5′ biotin–modified oligonucleotide probes that are individually synthesized and analyzed by electrospray ionization-mass spectrometry (ESI-MS) and optical density (OD) measurement. The probes are normalized before pooling to ensure that each probe is represented in the panel at the correct concentration. Probes that fail quality control are resynthesized. This manufacturing process gives the xGen Exome Hyb Panel v2 a unique advantage over array-derived pools in which missing or truncated probes cannot be identified before sequencing. IDT proprietary synthesis methods enable challenging probes—such as those with high GC and AT content—to be appropriately represented in the panel. The individual probes are synthesized on a larger scale than is used for array manufacturing, allowing IDT to produce a single, large lot of material that is aliquoted over time, providing consistent content from one aliquot to the next.
The xGen Exome Hyb Panel v2 consists of 415,115 probes that spans a 34 Mb target region (19,433 genes) of the human genome and 39 Mb of probe space—the genomic regions covered by probes. Our probes are designed using a new “capture-aware” algorithm and assessed with proprietary off-target analysis. All probes in the panel are manufactured under ISO 13485 standards, and then, mass spectrometry and dual quantification measurements of each probe are performed before they are pooled into the xGen Exome Hyb Panel v2. These measures ensure the quality of the probe and its appropriate representation in the final panel.
The xGen AML Cancer Hyb Panel is composed of individually synthesized and quality controlled xGen Hyb Probes that have a 5’ biotin group for capture with streptavidin-coated beads. The panel consists of 11,731 xGen Hyb Probes targeting empirically derived regions within 260 acute myeloid leukemia (AML)-associated genes.
Panel composition (target genes)
Table 1: Frequently researched genes included in the xGen AML Cancer Hyb Panel. *
| ASXL1 | FAM154B | IDH2 | KDR | NPM1 | PRAMEF2 |
| C17orf97 | FAM47A | JAK1 | KIT | NRAS | PTPN11 |
| CBL | FAM5C | JAK2 | KRAS | NTRK3 | RUNX1 |
| CEBPA | FLRT2 | JAK3 | LRRC4 | OR13H1 | TET2 |
| DNMT3A | FLT3 | KCNA4 | MLL3 | OR8B12 | TP53 |
| EGFR | GJB3 | KCNK13 | MYC | P2RY2 | TUBA3C |
| EZH2 | IDH1 | KDM6A | NF1 | PCDHB1 | WT1 |
* The panel does not target every exon in each gene, but targets all regions found to be mutated in the TCGA study [1].
Whole exome sequencing (WES) has traditionally been the standard method for research detecting a wide variety of inherited diseases. Now, IDT provides focused sequencing of specific genomic regions of interest to obtain deeper coverage for easier, more efficient, and identification of mutations in research samples. The xGen Inherited Diseases Hyb Panel targets disease-causing mutations as defined by The Human Gene Mutation Database (HGMD®) to combine the ease of WES with the low per sample cost of targeted panels.
The xGen Inherited Diseases Panel consists of 116,355 xGen Hyb Probes—spanning approximately 11.1 Mb of the human genome—and are designed specifically for research using targeted enrichment of genes and SNPs associated with the inherited diseases listed below. xGen Hyb Probes are individually-synthesized and quality-controlled 120 mer oligonucleotides, bearing a 5′ biotin modification and manufactured using our proprietary Ultramer™ synthesis technology.
Targeted disorders of the xGen Inherited Diseases Hyb Panel
- Autism spectrum disorders
- Cardiomyopathy
- Ciliopathies
- Congenital disorders of glycosylation (CDGs)
- Congenital myasthenic syndromes
- Epilepsy and seizure disorders
- Eye disorders
- Glycogen storage disorders
- Hearing loss
- Hereditary cancer syndrome
- Hereditary periodic fever syndromes
- Inflammatory bowel disease
- Lysosomal storage disorders
- Maturity onset diabetes of the young
- Multiple epiphyseal dysplasia
- Neuromuscular disorders
- Noonan syndrome and related disorders
- Peroxisome biogenesis disorders, Zellweger syndrome spectrum disorders
- Short stature panel
- Skeletal dysplasia
- X-linked intellectual disability
Every year, approximately 450 people in every 100,000 are diagnosed with cancer with a 5-year relative survival rate at 67.7%. Although survival rates have increased from less than 50% in 1975, there is a need for more basic research into the types of mutations and whether they are driver mutations or passenger mutations [1].
Research studies to produce a short list of mutated genes that are relevant to a multitude of cancer types, and that can be expanded to include additional cancer type–specific genes, would be invaluable in research. The Cancer Genome Atlas (TCGA) network performed a systematic analysis research study of more than 3,000 tumors from 12 cancer types to investigate underlying mechanisms of cancer initiation and progression and have identified 127 significantly mutated genes (SMGs) across these tumor types [2].
The xGen Pan-Cancer Hyb Panel is a hybridization capture panel based on the research findings of the TCGA network. The panel comprises xGen Hyb Probes, individually synthesized and quality controlled 120mer oligonucleotides bearing a 5′ biotin modification and manufactured using proprietary Ultramer™ synthesis technology.
Panel composition (target genes)
Table 1. List of genes that are associated with 12 different cancer types organized by gene pathway.
| Cellular process | Gene |
|---|---|
| Transcription factor/regulator |
VHL |
| Histone modifier |
MLL3 |
| Genome integrity |
TP53 |
| RTK signaling |
EGFR |
| Cell cycle |
CDKNA2 |
| MAPK signaling |
KRAS |
| PI(3)K signaling |
PIK3CA |
| TGF-β signaling |
SMAD4 |
| Wnt/β-catenin signaling |
APC |
| Histone |
HIST1H1C |
| Proteolysis |
FBXW7 |
| Splicing |
SF3B1 |
| HIPPO signaling |
CDH1 |
| DNA methylation |
DNMT3A |
| Metabolism |
IDH1 |
| NFE2L |
NEF2L2 |
| Protein phosphatase |
PPP2R1A |
| Ribosome |
RPL22 |
| TOR signaling |
MTOR |
| Other |
NAV3 |
IDT is committed to providing our customers with quality products for researchers working on the cutting edge of scientific discovery. The xGen CNV Backbone Hyb Panel was designed as part of the NGS Tech Access program at IDT, which is intended to accelerate innovation by enabling earlier access to our most advanced research tools. However, Tech Access products are not functionally QC-tested. These products are particularly well suited to experienced researchers who require the most up-to-date technology to unlock new discoveries.
Copy number variations (CNV)s are genomic changes that increase or decrease the number of copies of a particular gene or segment of gene. Underlying causes for these changes include duplications, inversions, translocations, and/or deletions.
The xGen CNV Backbone Hyb Panel has a modular formulation consisting of over 9,000 individually synthesized 5’-biotin modified oligonucleotide probes that are spaced approximately every 0.34 Mb across the human reference genome. After hybridization capture and next generation sequencing, the resulting data can be used for research on the root causes for cancer, autoimmune diseases, or studies on inherited diseases.
Hybridization capture panels, such as the xGen CNV Backbone Hyb Panel can identify the exact location of translocations, inversion, deletion, and/or duplication in the genome. Whether used with an IDT xGen Custom Hyb Panel, another predesigned hybridization panel, or as a standalone product, the results can identify variants in addition to CNV, such as single nucleotide variations (SNVs) and indels. Together, using the xGen CNV Backbone Hyb Panel provides an alternative way to understand the correlation of the human genome sequence and function.
Capturing 76 distinct, highly polymorphic sites, the xGen Human ID Hyb Panel enriches a definitive set of single nucleotide polymorphisms (SNPs), selected to identify a unique individual from a population of over 5 million biological research samples. The panel has been tested with DNA from formalin-fixed, paraffin-embedded (FFPE) tissues, cell-free DNA (cfDNA), and other research sample types.
The xGen Human ID Hyb Panel comprises 229 individually synthesized hybridization capture probes, each quality controlled by ESI mass spectrometry. The xGen Human ID Hyb Panel can be combined with any xGen Custom Hyb Panel as a spike-in panel in research studies. When used as a spike-in to other panels, the data had similar on-target percentages. The xGen Human ID Hyb Panel can also be run as a standalone† panel to confirm sample identity before performing larger, more expensive sequencing studies.
Use the xGen Human ID Hyb Panel to:
- Identify tumor/normal paired research samples
- Track and manage individual research samples from one experiment to the next
- Identify research sample misidentification
†The xGen Human ID Hyb Panel contains a relatively small number of probes. When used as a standalone panel, expect lower on-target capture results than when used as a spike-in to a larger panel. This is typical of small panels in hybridization capture enrichment.
The xGen Human Mitochondrial DNA (mtDNA) Hyb Panel provides coverage of the human mitochondrial genome in research studies. The panel consists of 138 individually-synthesized xGen Hyb Probes and delivers reliable coverage of the entire >16 kb mitochondrial genome.
This flexible, modular panel can be used as either a standalone† or spike-in panel, for cost-effective sequencing of variants in mitochondrial DNA in research samples. When added to other xGen Hyb Panels, the xGen Human mtDNA Hyb Panel does not affect the high target specificity or coverage uniformity of the original panel.
†Because the mitochondrial genome comprises a small target space, the xGen Human mtDNA Hyb Panel contains relatively few probes compared to other target capture panels. When used as a standalone panel, expect lower on-target results than when used as a spike-in to a larger panel. This is typical of small panels in hybridization capture enrichment.
The xGen Exome Hyb Panel v2 consists of 415,115 individually synthesized, 5’ biotinylated, oligonucleotide probes. Each probe was quality tested and combined so that the panel contains the correct concentration.
- Shown to have higher on-target rate than leading array based competitors since each probe is individually synthesized and then added to the panel
- Consistency across lots as each probe was synthesized on a large scale prior to panel creation
- High end-to-end coverage of the RefSeq genes even in areas of high GC content (refer to Figure 4)
xGen™ Exome Hyb Panel v2
Exome capture kit for target capture. 415,115 individually synthesized, quality controlled probes, manufactured to ISO standards, with deep, uniform coverage.
- 98% of genomic targets covered at greater than 0.2X mean coverage
- Detect variations
- Empirically derived targets
- Enjoy fast turnaround via easy online ordering and next day shipping
xGen™ AML Cancer Hyb Panel
Predesigned panel targeting >260 genes associated with AML for more efficient identification of disease-causing mutations.
- Obtain high coverage uniformity across all targets
- Identify variations reliably, with increased depth of coverage
- Expect fast turnaround time with easy online ordering and next-day shipping
xGen™ Inherited Diseases Hyb Panel
Predesigned panel for targeted enrichment of genes and SNPs associated with inherited diseases.
- Obtain high coverage uniformity across all targets
- Identify variations reliably with increased depth of coverage
- Enjoy fast turnaround (TAT) via easy online ordering and next-day shipping
xGen™ Pan-Cancer Hyb Panel
Predesigned panel that captures 127 significantly mutated genes implicated across 12 tumor types for deeper sequencing coverage.
- Identify changes in copy number throughout the human genome
- Use as either a spike-in or a standalone panel
- Enjoy fast turnaround via easy online ordering and next day shipping
xGen™ CNV Backbone Hyb Panel
Predesigned spike-in panel that can be used to identify copy number variations in human NGS data.
- Trace NGS sample identity during data analysis
- Use as a spike-in or standalone panel with unique, modular format
- Consists of individually synthesized and quality-controlled 5’ biotin-modified hybridization capture probes
- Compatible with FFPE and cfDNA DNA samples
xGen™ Human ID Hyb Panel
Predesigned spike-in panel for targeted enrichment of a definitive set of single nucleotide polymorphisms (SNPs), selected to identify a unique individual from a population of over 5 million biological samples.
- Identify important variants in research samples with complete coverage of the human mitochondrial genome
- Easily expand your target space with a predesigned spike-in panel that can be used together with other target enrichment panels
- Consistently achieve reliable results with individually synthesized probes
xGen™ Human mtDNA Hyb Panel
Predesigned spike-in panel for cost-effective and accurate identification of sequence variants in mitochondrial DNA.
- Identify known and novel mutations arising from diverse strains
- Available as a bundle with xGen™ Universal Blockers-TS Mix and xGen Hybridization and Wash Kit
xGen™ SARS-CoV-2 Hyb Panel
Hybridization capture probes designed to create SARS-CoV-2 sequencing libraries.
 Forgalmazott termékeink gyártói - keressen gyártó szerint a logóra kattintva
Forgalmazott termékeink gyártói - keressen gyártó szerint a logóra kattintva