xGen Methyl-Seq Library Prep Kit utilizes Adaptase™ technology that efficiently captures bisulfite-converted ssDNA molecules into library molecules for epigenetic research studies. The resultant libraries represent uniform and comprehensive genome coverage.
xGen NGS—made reliable.
-
IDT termékek szállítási költségei 2024.04.-től:
A szintézis helyétől, valamint a szállítási körülményektől függően az alábbi költségek kerülnek felszámításra:
-
Belgiumból: nettó 6 900 Ft
USA-ból: nettó 34 000 Ft
Belgiumból, szárazjégen: nettó 37 000 Ft
USA-ból, szárazjégen: nettó 45 000 Ft
- Oldatok szállítási költsége: mennyiség és súly függvénye, kérjen ajánlatot a Bio-Science Kft.-től!
Általános tájékoztató az IDT USA gyártási központjából érkező „custom” termékcsoportokról:
szintetikus biológiai termékek:
- Gének
- gBlocks, eBlocks
- Ultramerek
- RNS-oligók
- CRISPR guide RNS-ek
- oPools
qPCR próbák:
a szintézis helye a szekvencia komplexitásától, valamint a választott festék/ quencher kombinációtól függ
szolgáltatások:
- PAGE tisztított termékek
stock termékek:
nem áll teljes lista rendelkezésre, kérjen ajánlatot a Bio-Science Kft.-től!
-
- Comprehensive sample representation—uniform methylome genome coverage by whole genome bisulfite sequencing (WGBS)
- Low bias library preparation from low input amounts—supports inputs down to single cell
- High multiplex capability—pre-plated UDI primers enable multiplexing of up to 1536 samples
- Compatible with xGen Normalase™ technology—streamlines normalization of libraries enzymatically for a single library pool
- Powered by xGen Adaptase technology—template independent adapter attachment reduces workflow time while generating a less biased library
- Simple, 2-hour workflow
- Workflow design for easy automation
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend for these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.
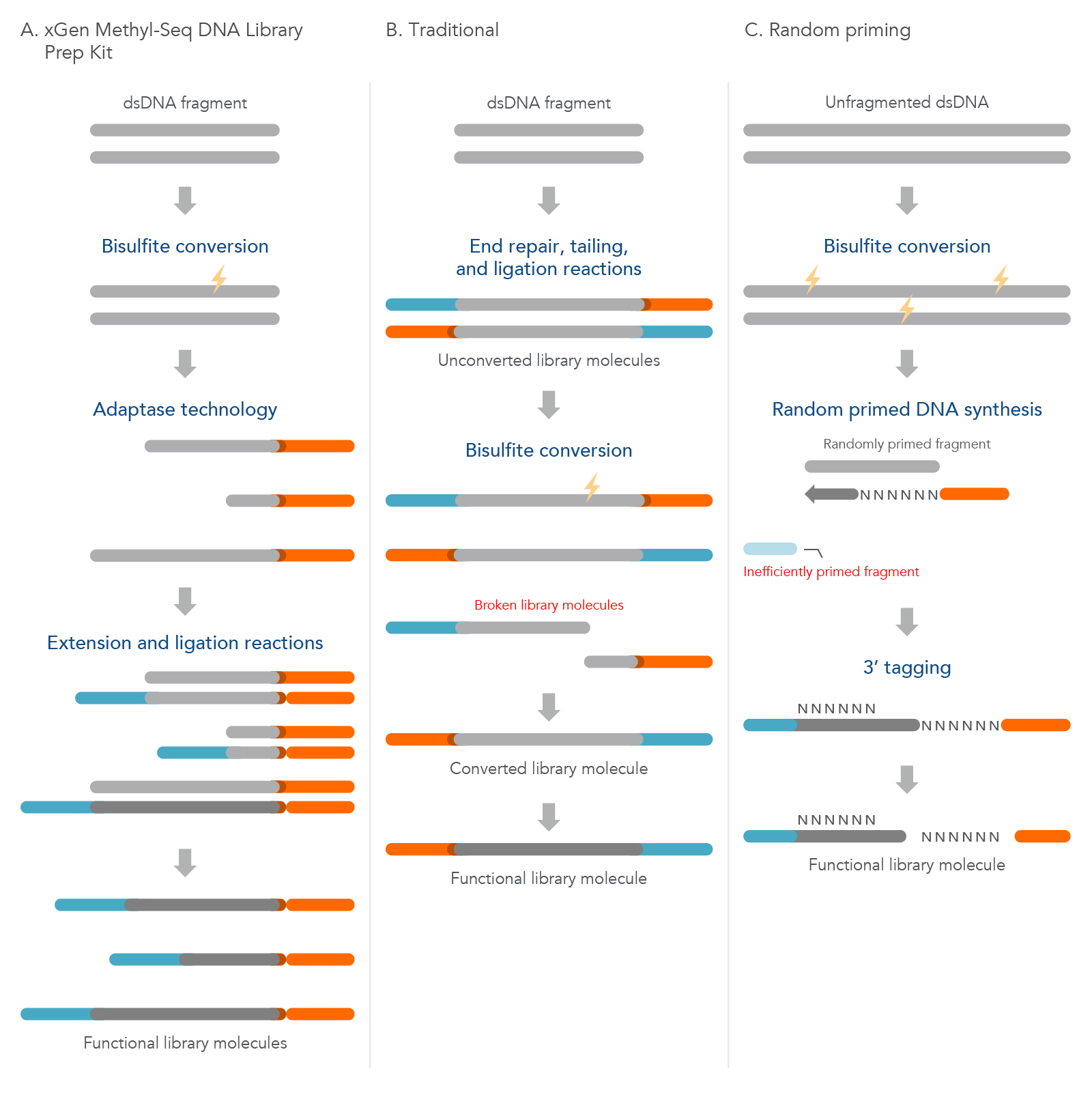
Methyl-seq library preparation workflow comparisons
The xGen Methyl-Seq DNA Library Prep Kit has a post-bisulfite library preparation workflow to maximize library complexity by converting bisulfite-induced single-stranded fragments directly into library molecules. Traditional methods construct libraries from dsDNA using methylated adapters followed by bisulfite conversion that leads to significant library loss due to bisulfite-induced DNA fragmentation. The random priming methods are also post-bisulfite but have reduced library complexity due to a lower efficiency biased workflow (see Table 2; Figure 3).
Figure 3. Comparison of the xGen Methyl-Seq DNA Library Prep Kit workflow to two other common methods. (A) Before using the xGen Methyl-Seq DNA Library Prep Kit, double-stranded DNA (dsDNA) fragments undergo bisulfite conversion. The kit then directly converts these fragments into library molecules as described previously. (B) The traditional method of methylation sequencing builds the library before bisulfite conversion, which can lead to broken library molecules. (C) The random priming method converts unfragmented dsDNA with bisulfite treatment first, but due to inefficient random priming, many library molecules are lost and are not represented in the final library.
Library complexity and coverage from 1 ng of input DNA
To demonstrate uniform and comprehensive genome coverage using the xGen Methyl-Seq DNA Library Prep Kit, a titration experiment using 100 ng, 10 ng, or 1 ng of Arabidopsis thaliana genomic DNA was performed in duplicate and compared to two alternative methods (random primer and traditional) using these same input DNA samples. Each A. thaliana file was normalized to 30.2 million reads and data reported as an average of duplicate bisulfite-converted samples.
The xGen Methyl-Seq DNA Library Prep Kit demonstrates a higher, more comprehensive genome coverage and lower duplication rates than both alternative methods for each of the input samples—even the lowest concentration of 1 ng Arabidopsis DNA. The traditional method had equivalent results at the 100 ng input but lost significant complexity with the lower inputs. In contrast, the random-primed method had lower complexity and higher duplicate reads for each input concentration.
Table 2. Comparison of NGS library metrics obtained using the xGen Methyl-Seq DNA Library Prep Kit to traditional or random-primed methylation sequencing workflows.
| Input | Sample | % Reads aligned | Genome coverage | % Duplicate reads | Est. library size (Millions) | % CpX missing | % CpX covered ≥ 10X |
|---|---|---|---|---|---|---|---|
| 100 ng Arabidopsis | xGen Methyl-Seq | 89.6 | 22.0X | 1.9 | 714 | 0.56 | 92.2 |
| Traditional | 80.2 | 21.0X | 2.7 | 604 | 0.57 | 88.1 | |
| Random Primer | 71.4 | 16.0X | 22.1 | 48 | 7.70 | 39.4 | |
| 10 ng Arabidopsis | xGen Methyl-Seq | 87.8 | 22.0X | 2.7 | 406 | 0.58 | 90.4 |
| Traditional | 76.7 | 19.0X | 11.9 | 70 | 0.57 | 83.9 | |
| Random Primer | 71.9 | 16.0X | 22.2 | 45 | 5.2 | 45.2 | |
| 1 ng Arabidopsis | xGen Methyl-Seq | 83.3 | 18.0X | 18.2 | 38 | 0.59 | 77.1 |
| Traditional | 80.7 | 10.0X | 62.3 | 6 | 2.00 | 17.0 | |
| Random Primer | 73.4 | 12.0X | 46.1 | 12 | 6.60 | 31.3 | |
| 10 ng Human | xGen Methyl-Seq | 86.4 | 8.9X | 7.9 | 1,393 | N/A | N/A |
DNA methylation is the most widely studied epigenetic modification. DNA methylation occurs primarily on cytosine bases in the CpG dinucleotide motif in most mammalian cells, and across all cytosine contexts in plant genomes. DNA methylation is often associated with the regulation of gene expression through its presence or absence in CpG islands among gene promoter regions, and these methylation patterns can be used to identify different cell types and disease states [1].
IDT offers two products for methylation sequencing:
- xGen Methyl-Seq DNA Library Prep Kit for whole genome and targeted methylation sequencing
- xGen Adaptase Module for single cell methylation sequencing
Some common applications of methyl-seq include:
- Whole genome bisulfite sequencing (WGBS) research
- Identifying genome-wide methylation of cell-free (cfDNA) in research studies
- Hybridization capture for targeted methylation studies
- Bisulfite-converted DNA enriched by MeDIO, chromatin immunoprecipitation (ChIP), or other research methods
xGen Methyl-Seq Library Prep Kits
The xGen Methyl-Seq DNA Library Prep Kit can be used for WGBS and targeted bisulfite sequencing with the addition of an xGen Custom Hyb Panel. The kit’s post-bisulfite library preparation workflow offers an efficient library preparation method. The kit is compatible with low input quantities such as cfDNA for biomarker research studies. The xGen Methyl-Seq DNA Library Prep Kit is also compatible with bisulfite-converted DNA samples enriched by ChIP or other methods as well as ancient DNA samples that may be deaminated.
2-hour workflow for bisulfite-converted samples
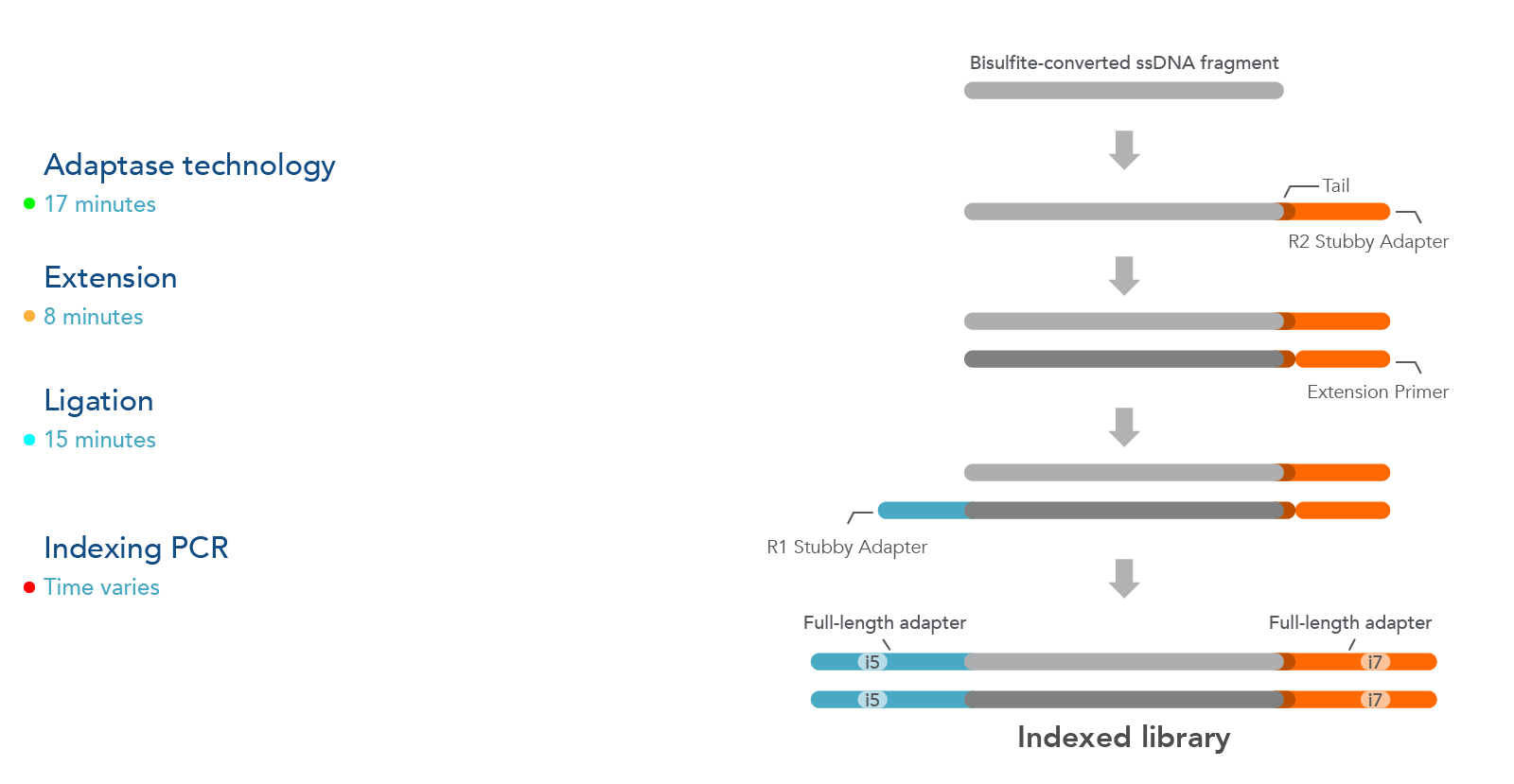
The xGen Methyl-Seq DNA Library Prep Kit workflow maximizes DNA recovery through a post-bisulfite library preparation, utilizing an efficient adapter attachment that is compatible with single-stranded, bisulfite-converted DNA (Figure 1). Library complexity from this kit is significantly greater than those from methods that bisulfite convert after library construction (see Table 2).
Additionally, the template-independent adapter attachment chemistry of the xGen Methyl-Seq DNA Library Prep Kit provides a more complete, less biased library as observed from comprehensive methylome coverage by WGBS (see Table 2).
Figure 1. The xGen Methyl-Seq DNA Library Prep Kit workflow constructs libraries from single-stranded, bisulfite-converted DNA fragments. The Adaptase step simultaneously performs end repair, tailing, and ligation of the R2 Stubby Adapter to the 3’ end of each fragment. The extension step produces a uracil-free strand and the ligation step adds R1 Stubby Adapter to the uracil-free strand. Indexing PCR increases library yield and incorporates full-length adapters with sample-specific index sequences.
Table 1. Specifications and features of the xGen Methyl-Seq DNA Library Prep Kit.
| Feature | xGen Methyl-Seq DNA Library Prep Kit |
|---|---|
| Sample types | gDNA, FFPE, cfDNA |
| Input DNA amount | 100 pg to 100 ng |
| Indexing compatibility | CDI primers up to 96-plex, with or without Normalase technology UDI primers up to 1536-plex, with or without Normalase technology |
| System compatibility and multiplexing format | Illumina® sequencing instruments |
| Workflow compatibility | Manual & automated. For list of liquid handlers and scripts, please inquire. |
xGen Adaptase Module for single cell methylation sequencing
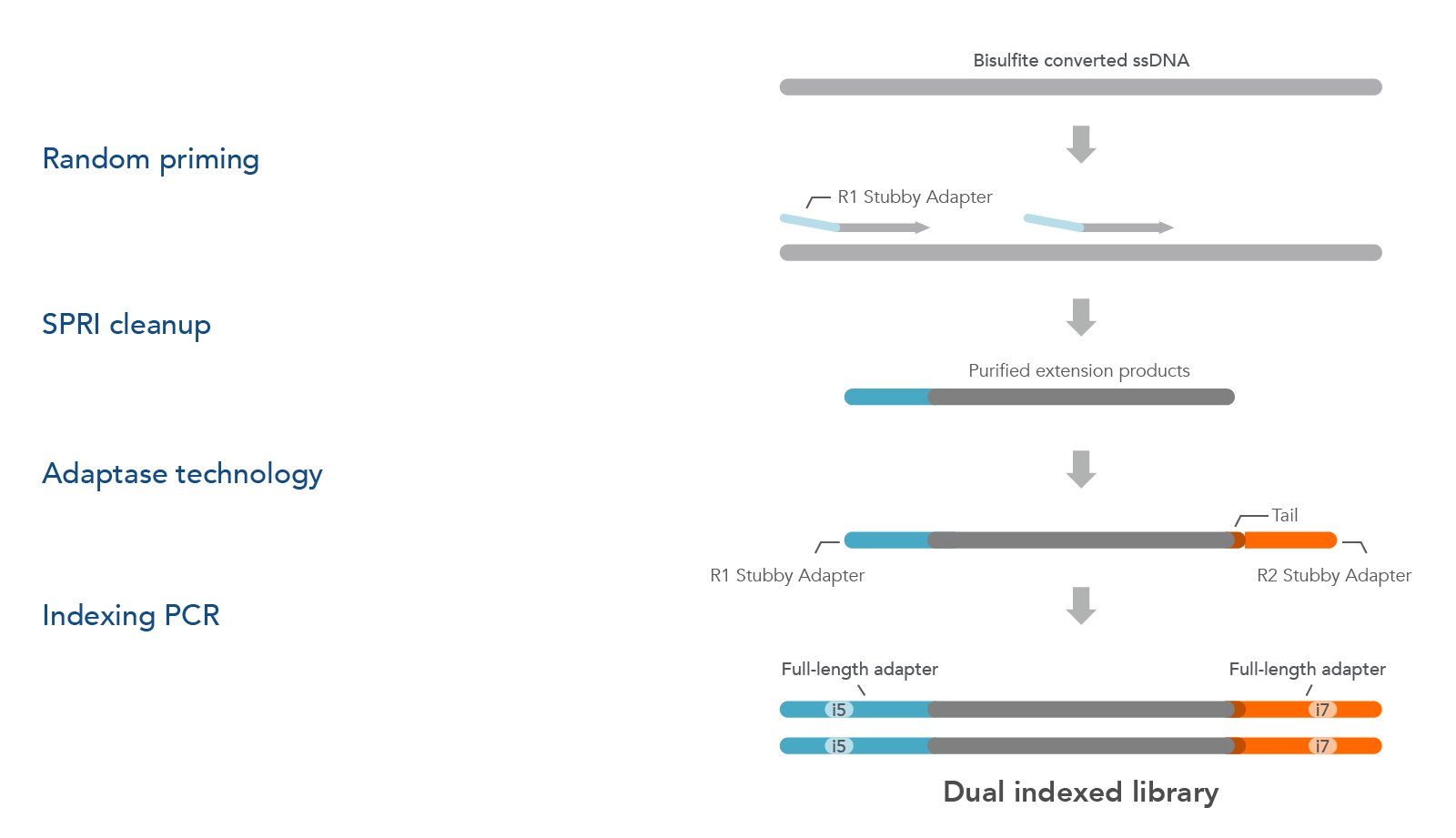
xGen Adaptase Module is also available to buy as a separate product (see ordering section). The xGen Adaptase Module supports a workflow that constructs NGS libraries from bisulfite-converted DNA from single cells. The xGen Adaptase Module maximizes the recovery of low concentrations of single-stranded DNA. In internal experiments, libraries created with this module have consistently exhibited higher complexity with reduced composition bias to provide a more faithful representation of the methylome. Using the in-line barcoding option, cells can be multiplexed to increase throughput and further reduce library prep costs.
The Adaptase Module only supplies the Adaptase reagents for the workflow (Figure 2). Random priming, purification, and indexing PCR reagents must be sourced separately. The Adaptase Module can be used for many applications, such as cataloging cell populations within heterogeneous tissues, assessing normal tissue for regulation of cellular mechanisms (e.g., differentiation), gaining insight into epigenetic alterations in disease states, and exploring across species to identify evolutionary conservation of epigenomic regulation.
Figure 2. The xGen Adaptase Module for single-cell bisulfite sequencing library preparation. First, bisulfite-converted ssDNA fragments are copied using custom random primers that are designed to incorporate the R1 Stubby Adapter (light orange) onto the 5´ end of the extension products using commercially available enzymes and buffers. Random primers can be designed to incorporate in-line barcode sequences for downstream multiplexing. During the next step, the proprietary xGen Adaptase reagent simultaneously tails and ligates the R2 Stubby Adapter (light blue) to the 3´end of the fragments. To complete the library, library amplification with indexing primers (not part of the xGen Adaptase Module) creates dual indexed library fragments for Illumina sequencing instruments.
NOTE: To support the workflow above, custom oligonucleotide primers and commercially available enzymes and buffers for complete functionality are sourced separately. Please see the xGen Adaptase Module for Single-Cell Methyl-Seq Library Preparation protocol for a complete list of required materials.
For product data on the individual xGen Adaptase Module, please review [2] or [3]
xGen™ Methyl-Seq DNA Library Prep Kit
Library prep kit for whole genome bisulfite sequencing or targeted Methyl-Seq; Adaptase-based chemistry for library prep of single stranded bisulfite converted DNA, includes amplification polymerase, bisulfite conversion module not provided (recommend Zymo), indexing primers also supplied separately.
xGen™ Adaptase™ Module
Adaptase reagents for adding a stubby adapter to ssDNA substrates (for single cell Methyl-Seq workflow, not a complete library prep).
xGen™ Normalase™ Module
Core kit for performing Normalase, includes Normalase I and Normalase II reagents and Normalase terminal primers, Normalase indexing primers sold separately.
xGen™ Normalase™ UDI Primers
Premixed, unique dual index primers designed for Normalase™ workflow. Individual plates of 96 indexes or sets of 384 indexes are available. Or choose all four sets for a total of 1536 index pairs. All plates are single-use to minimize the risk of contamination by multiple handling events.
xGen™ UDI Primer Pairs
Premixed unique dual index primer pairs for sample indexing by PCR. These are delivered as single-use plates containing either 16 or 96 primer pairs, or for high-volume/high-throughput customers we have bulk 2 nmol offerings of primer with 10 nt indexes. The 10 nt indexes are available in sets of 384, 768, and 1536 index pairs. All plates except the bulk 2 nmol offerings are single-use to minimize the risk of contamination by multiple handling events.
xGen™ Input DNA Quantification Primers
Two primer pairs to human ALU repeat sequences to determine usable input DNA, assesses DNA quantity and integrity with ratio of 115/247 bp amplicons; comprises primer pairs only; qPCR Master Mix sourced separately (separate vendor).
Legacy Swift Indexing Primers
Legacy Swift Primers, CDI and UDI indexing strategies. The xGen™ CDI Primers are individual tubes of indexes, to be arrayed in a combinatorial fashion for up to 96 possible barcode combinations, and the xGen™ UDI Primers, Plate 2 are premixed Unique Dual Index primers in single-use plates. Both products have 8nt barcodes. Note that these primers are not compatible with any other IDT indexing primers, due to potential demultiplexing issues.
xGen™ Normalase™ CDI Primers
Combinatorial dual index primers designed for Normalase workflow. Consists of 20 individual tubes of indexes that support up to 96 combinations of i7-i5 indexes.
 Forgalmazott termékeink gyártói - keressen gyártó szerint a logóra kattintva
Forgalmazott termékeink gyártói - keressen gyártó szerint a logóra kattintva