xGen Hybridization Capture Core Reagents provide reliable targeted sequencing functionality across a range of panel sizes and multiplexing levels. They work with xGen Hyb Panels for a complete, high quality target enrichment solution.
xGen NGS—made to maximize.
FIGYELEM!
IDT termékek szállítási költségei 2024-től:
A szintézis helyétől, valamint a szállítási körülményektől függően az alábbi költségek kerülnek felszámításra:
-
Belgiumból: nettó 6 900 Ft
USA-ból: nettó 34 000 Ft
Belgiumból, szárazjégen: nettó 37 000 Ft
USA-ból, szárazjégen: nettó 45 000 Ft
- Oldatok szállítási költsége: mennyiség és súly függvénye, kérjen ajánlatot a Bio-Science Kft.-től!
Általános tájékoztató az IDT USA gyártási központjából érkező „custom” termékcsoportokról:
szintetikus biológiai termékek:
- Gének
- gBlocks, eBlocks
- Ultramerek
- RNS-oligók
- CRISPR guide RNS-ek
- oPools
qPCR próbák:
a szintézis helye a szekvencia komplexitásától, valamint a választott festék/ quencher kombinációtól függ
szolgáltatások:
- PAGE tisztított termékek
stock termékek:
nem áll teljes lista rendelkezésre, kérjen ajánlatot a Bio-Science Kft.-től!
The xGen™ Hybridization and Wash v2 Kit is designed for use with xGen Hyb Panels and xGen Universal Blockers. This kit consists of two core components—the xGen Hybridization & Wash v2 Reagents and the xGen Hybridization & Wash v2 Beads—to perform the hybridization capture workflow. The latest version of the kit contains a new, internally developed, xGen 2x HiFi PCR Master Mix and is compatible with the protocol, xGen hybridization capture of DNA libraries for NGS target enrichment, to provide a complete, high-quality hybridization capture solution for targeted NGS research.
Prevent adapter cross-hybridization by using xGen Universal Blockers in your hybridization capture reactions. Adapter sequences are ligated to all library fragments, both on-target and off-target before enrichment. These adapter sequences can hybridize with each other during enrichment, creating a "daisy-chain" effect, with off-target fragments being captured alongside on-target fragments, which can impact sequencing efficiency.
xGen Universal Blockers bind to platform adapter sequences on a designated strand (usually the inverse of the synthetic adapter) to help prevent non-specific binding. Blocking the adapter sequences significantly improves capture results [1, 2].
Automation
We have partnered with several instrument vendors to develop automation scripts for research using the xGen hybridization capture protocol on liquid handling robots. Scripts are currently available for the following:
- Beckman Coulter Biomek™ i5 and i7 Automated Workstations
- Hamilton NGS STAR™ for Library Prep Workstation
- Perkin Elmer Sciclone™ Workstation
- Achieve uniform coverage and reliable capture functionality across a range of xGen Hyb Panels
- Generate targeted NGS data with a quick and easy workflow
- Updated, automation-friendly protocol enables various levels of throughput
- xGen Universal Blockers effectively block a variety of index adapter designs with proprietary sequence modifications improving on-target results
- xGen 2x HiFi PCR Mix now included, so no need to source components from multiple vendors
-
xGen Hybridization and Wash v2 Kit - 16 rxn
Optimized buffers, Cot DNA, xGen 2x HiFi PCR Mix, and streptavidin beads for use with xGen™ Hyb Panels in hybridization capture target enrichment. This kit includes two products - the reagents component and the streptavidin beads component, used together for performing hybridization target enrichment. Both components must be purchased together in equal quantities.
xGen Hybridization and Wash v2 Kit - 96 rxn
Optimized buffers, Cot DNA, xGen 2x HiFi PCR Mix, and streptavidin beads for use with xGen™ Hyb Panels in hybridization capture target enrichment. This kit includes two products - the reagents component and the streptavidin beads component, used together for performing hybridization target enrichment. Both components must be purchased together in equal quantities.
xGen™ Universal Blockers
Proprietary blockers for dual- and single-index adapters, to reduce non-specific adapter interaction during probe hybridization and increase on-target capture performance.
xGen™ Library Amplification Primer Mix
Premixed P5/P7 primer pairs to amplify indexed NGS libraries, to achieve the necessary yield for sequencing. Available in three reaction sizes, for flexibility.
xGen™ Human Cot DNA
Placental DNA enriched for repetitive DNA seq
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend for these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.
Achieve uniform sequence coverage using automation, as compared to manual (tube and plate).
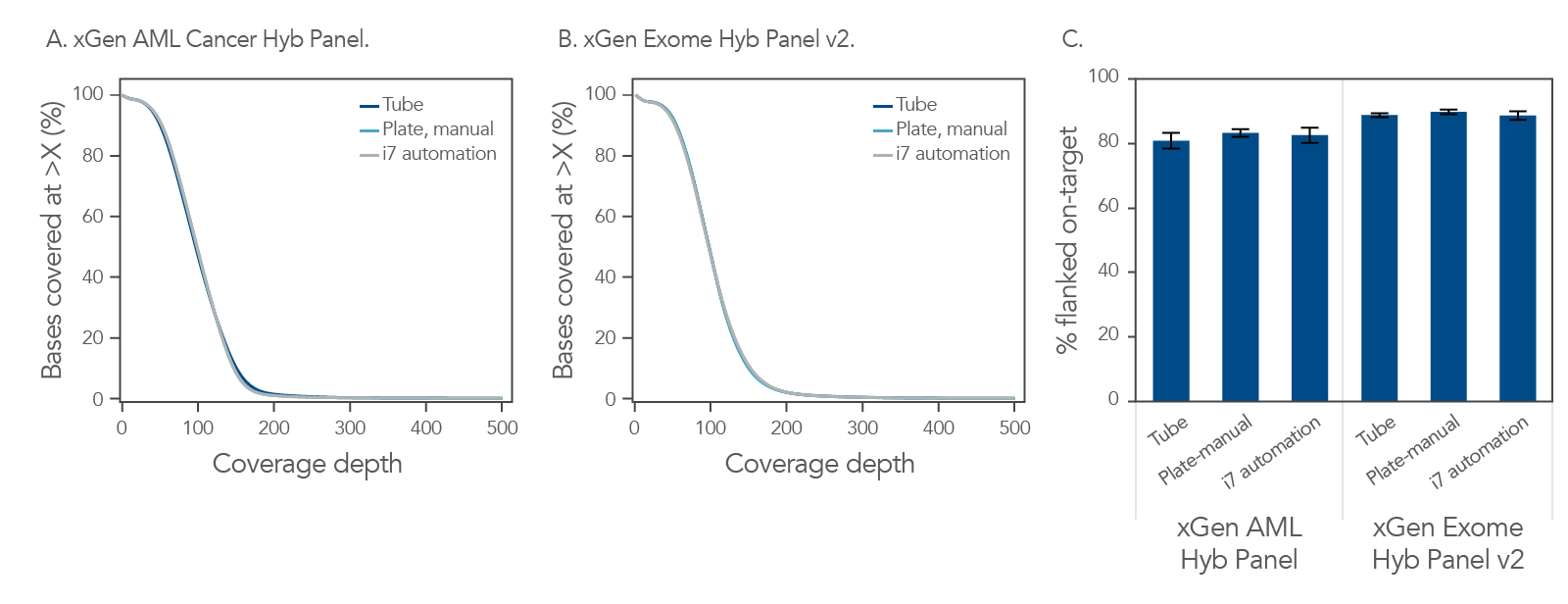
Figure 1. Consistent coverage independent of workflow format and type of hyb panel. All libraries were prepared on the Biomek i-series i7 automation platform, using the xGen cfDNA & FFPE DNA Library Prep v2 MC Kit (n = 96, avg shear size of ~150 bp) with 100 ng of Coriell NA24385 input DNA. The DNA libraries were subsequently enriched using either the xGen AML Cancer Hyb Panel (A) or xGen Exome Hyb Panel v2 (B), with the xGen Hybridization and Wash v2 Kit and xGen Universal Blockers. Library prep was done using the indicated combinations of methods (manual or automated) and formats (plate or tubes). (A) Hyb capture with the xGen AML Cancer Hyb Panel provided equivalent coverage for manual or automated in plate or tubes workflows, n = 1 for each test. (B) Equivalent coverage was obtained for manual versus automated and plates versus tubes for libraries captured with the xGen Exome Hyb Panel v2, n =1 for each test. (C) Consistent % on-target for each method and for each panel. For the different methods, replicate capture samples for each panel in single-plex, n = 18 for automation, n = 16 for manual plate, and n = 8 for tube were used. Automated capture was also performed using the Biomek i-series i7 automation platform with automation scripts developed jointly by IDT and Beckman Coulter Life Sciences according to the xGen hybridization capture of DNA libraries for NGS target enrichment protocol. Sequencing was performed on a NextSeq™ 2000 instrument to generate 2x150 bp, paired-end reads. In summary, coverage is consistent regardless of workflow method, when using the xGen Hybridization and Wash v2 Kit. Both the libraries and captures were amplified using the proprietary xGen 2x HiFi PCR Master Mix, now included in the xGen Hybridization & Wash v2 Reagents pack.
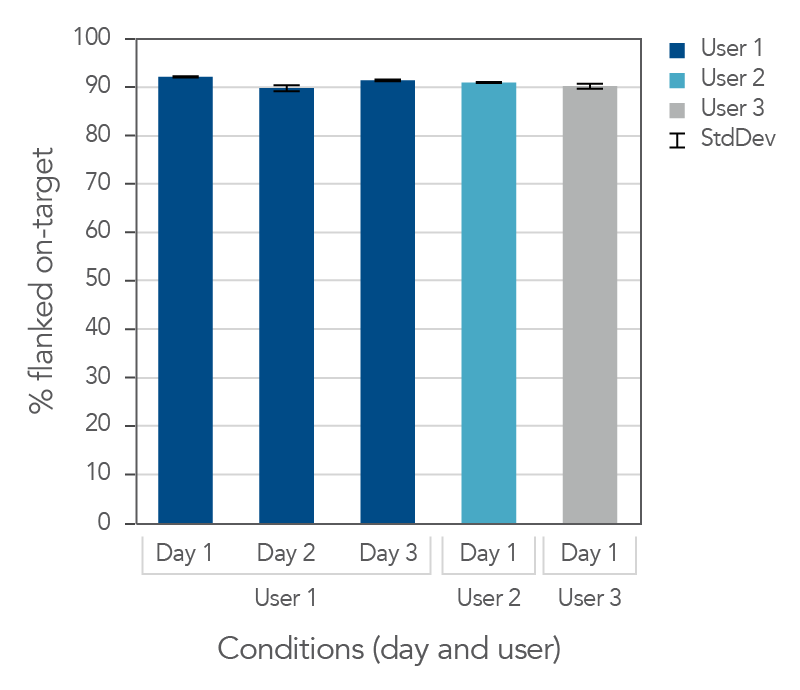
Consistent on-target rates between users and across days
Figure 2. Day-to-day repeatability and user-to-user reproducibility of on-target rates for target enrichment in 96-well plates. DNA libraries (n = 4) were prepared from 25 ng mechanically sheared NA12878 gDNA (Coriell) using the xGen cfDNA & FFPE DNA Library Prep v2 MC Kit. Libraries were then hybridized overnight using the xGen AML Cancer Hyb Panel and captured with the xGen Hybridization and Wash v2 Kit, and xGen Universal Blockers. Samples were sequenced on a NextSeq 500 (IIIumina) by paired end reads to an average read depth of 20 million read pairs and subsampled to 15 million read pairs. To test user-to-user reproducibility, target capture was performed by multiple users, using the manual protocol for 96-well plates. To test day-to-day repeatability, User 1 performed target capture on three separate days. On-target values (with 150 bp flank) were averaged for each data set. Both the libraries and captures were amplified using the novel, internally created, xGen 2x HiFi PCR Mix, now included in the xGen Hybridization & Wash v2 Reagents pack.
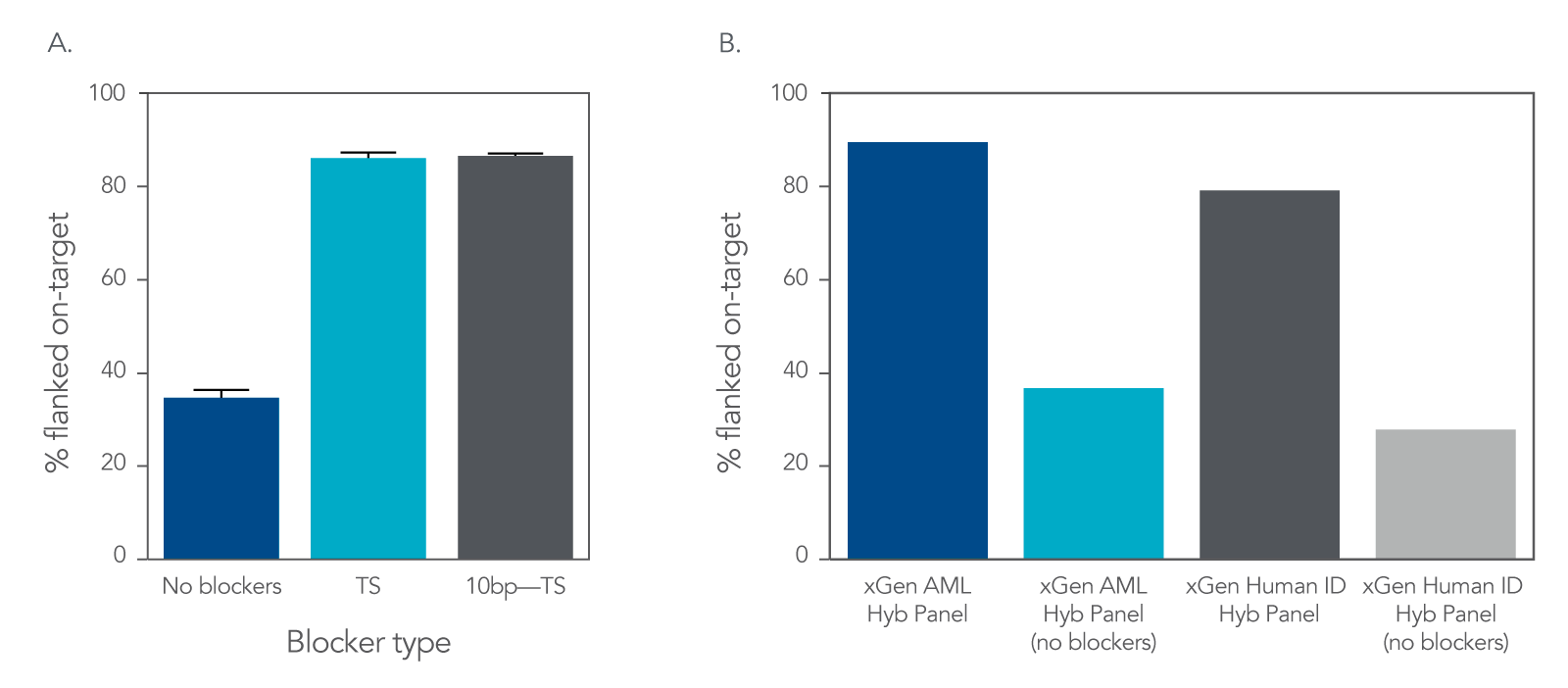
Increase on-target rates by using xGen Universal Blockers
Figure 3. An example of improved on-target rate delivered by xGen Universal Blockers across Illumina TruSeq® (A) and Illumina Nextera® (B) adapter styles. (A) 1 µg input DNA libraries were prepared from human genomic cell line NA12878 (Coriell) using either 8 nt or 10 nt adapters and enriched using the xGen AML Cancer Hyb Panel (single-plex, n = 3 for each condition) with xGen™ Universal Blockers TS, xGen™ Universal Blockers 10bp TS or without blockers. Sequencing was performed on a NextSeq 500 System (Illumina) to generate 2x150 bp, paired-end reads. (B) 50 ng cell line NA12878 (Coriell) gDNA was used for Nextera library preparation. Amplified libraries were enriched in 4-plex reactions (n = 3) using the xGen AML Cancer Hyb Panel, or in 12-plex (n = 1) using the xGen Human ID Hyb Panel, with xGen Universal Blockers NXT or without blockers (n = 1). Sequencing was performed on a NextSeq System (Illumina) to generate 2x75 bp paired-end reads. Libraries and captures from both figures were amplified using KAPA 2x HiFi PCR Mix, as this data was generated prior to release of xGen 2x HiFi PCR Mix.
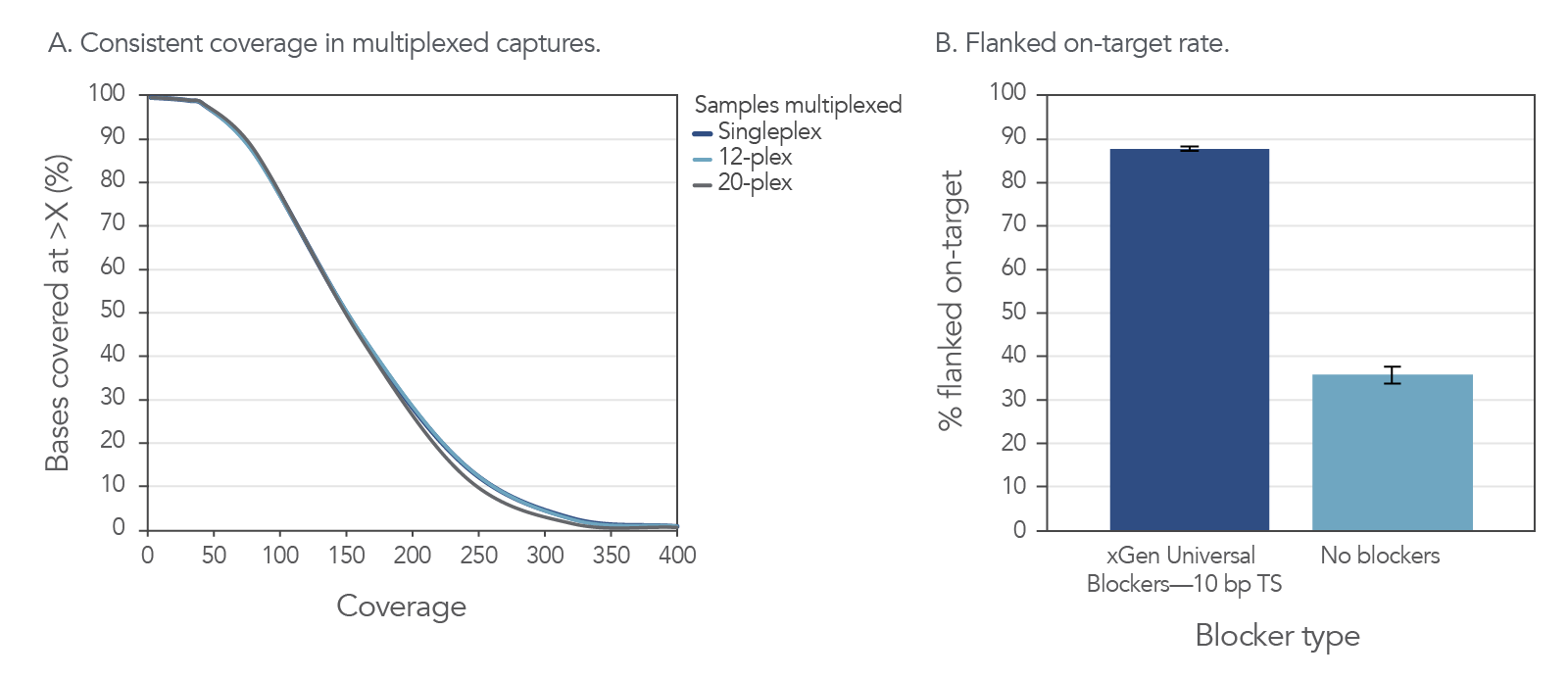
High on-target and uniform coverage with xGen Universal Blockers 10bp TS
Figure 4. Consistent coverage in multiplexed captures and improved on-target rates with the addition of xGen Universal Blockers. (A) Example of consistent results with multiplexed samples using the xGen Universal Blockers 10bp TS. DNA libraries (500 ng input per sample) from cell line NA12878 (Coriell) were enriched in single (n = 8) or multiplex reactions (n = 3) using the xGen AML Cancer Hyb Panel. The libraries were created using 10 bp TruSeq adapters with unique dual indexes. Sequencing was performed on a NextSeq 500 System (Illumina) with 2x150 bp paired-end reads. Coverage was consistent across all multiplexing levels tested. (B) Example of improved on-target rates delivered by xGen Universal Blockers 10bp TS. DNA libraries (500 ng input per sample) from cell line NA12878 were enriched in single-plex reactions (n = 3) using the xGen AML Cancer Hyb Panel with and without xGen Universal Blockers 10bp TS. Sequencing was performed on a NextSeq 500 System to generate 2x150 bp, paired-end reads. On-target values (with 150 bp flank) were averaged across experiments. Libraries and captures from both figures were amplified using KAPA 2x HiFi PCR Mix, as this data was generated prior to release of xGen 2x HiFi PCR Mix.
- Achieve uniform coverage and reliable capture functionality across a range of xGen Hyb Panels
- Generate targeted NGS data with a quick and easy workflow
- Updated, automation-friendly protocol enables various levels of throughput
- xGen Universal Blockers effectively block a variety of index adapter designs with proprietary sequence modifications improving on-target results
- xGen 2x HiFi PCR Mix now included, so no need to source components from multiple vendors
-
xGen Hybridization and Wash v2 Kit - 16 rxn
Optimized buffers, Cot DNA, xGen 2x HiFi PCR Mix, and streptavidin beads for use with xGen™ Hyb Panels in hybridization capture target enrichment. This kit includes two products - the reagents component and the streptavidin beads component, used together for performing hybridization target enrichment. Both components must be purchased together in equal quantities.
xGen Hybridization and Wash v2 Kit - 96 rxn
Optimized buffers, Cot DNA, xGen 2x HiFi PCR Mix, and streptavidin beads for use with xGen™ Hyb Panels in hybridization capture target enrichment. This kit includes two products - the reagents component and the streptavidin beads component, used together for performing hybridization target enrichment. Both components must be purchased together in equal quantities.
xGen™ Universal Blockers
Proprietary blockers for dual- and single-index adapters, to reduce non-specific adapter interaction during probe hybridization and increase on-target capture performance.
xGen™ Library Amplification Primer Mix
Premixed P5/P7 primer pairs to amplify indexed NGS libraries, to achieve the necessary yield for sequencing. Available in three reaction sizes, for flexibility.
xGen™ Human Cot DNA
Placental DNA enriched for repetitive DNA sequences. Used to block nonspecific binding in DNA hybridization reactions.
 Forgalmazott termékeink gyártói - keressen gyártó szerint a logóra kattintva
Forgalmazott termékeink gyártói - keressen gyártó szerint a logóra kattintva