xGen NGS—ready to react.
IDT termékek szállítási költségei 2024.04.-től:
A szintézis helyétől, valamint a szállítási körülményektől függően az alábbi költségek kerülnek felszámításra:
-
Belgiumból: nettó 6 900 Ft
-
USA-ból: nettó 34 000 Ft
-
Belgiumból, szárazjégen: nettó 37 000 Ft
USA-ból, szárazjégen: nettó 45 000 Ft
- Oldatok szállítási költsége: mennyiség és súly függvénye, kérjen ajánlatot a Bio-Science Kft.-től!
Általános tájékoztató az IDT USA gyártási központjából érkező „custom” termékcsoportokról:
szintetikus biológiai termékek:
- Gének
- gBlocks, eBlocks
- Ultramerek
- RNS-oligók
- CRISPR guide RNS-ek
- oPools
qPCR próbák:
a szintézis helye a szekvencia komplexitásától, valamint a választott festék/ quencher kombinációtól függ
szolgáltatások:
- PAGE tisztított termékek
stock termékek:
nem áll teljes lista rendelkezésre, kérjen ajánlatot a Bio-Science Kft.-től!
xGen™ Oncology & Inherited Diseases Amplicon Panels
Compatible with FFPE and cell-free DNA samples
Confident variant identification as low as 1% allele frequency facilitates research on tumor growth and identification of driver mutations in a fast, easy, single-tube workflow.
xGen NGS—made for oncology and inherited disease research.
xGen™ Metagenomics Amplicon Panels
Research identities in mixed microbial community
16S v2 and ITS1 panels facilitate NGS research on complex microbial communities using a single primer pool targeting the 16S rRNA gene or ITS1 region. These panels can be customized with additional targets including antibiotic resistance or virulence genes.
xGen NGS—made for metagenomics.
xGen™ SARS-CoV-2 Amplicon Panels
Targeted sequencing to research SARS-CoV-2 viral genomics
Overlapping primers produce 99.7%† genomic coverage of SARS-CoV-2 lineages. These consistent primers obtain sequence data from viral concentrations as low as 10–100 viral copies without the need to evaluate new primer sets for emerging lineages.
xGen NGS—made for COVID-19 research.
xGen™ Monkeypox Virus Amplicon Panel
xGen Amplicon Technology uses overlapping primer sets to provide comprehensive coverage of positions 6760–190,905 (ITRs not included) from as few as 300 monkeypox viral genome copies. Track the spread in a single-tube, ~2.5-hour workflow.
xGen NGS—made for monkeypox research.
xGen™ Sample Identification (ID) Amplicon Panel
Track and manage samples, including matched pairs
The power of discrimination of this amplicon panel is over 1 in 85,000, making it suitable for longitudinal research studies and scenarios in which genetic fingerprinting is relevant to research design and analysis.
xGen NGS—made for sample identification research.
ARTIC SARS-CoV-2 Amplicon Panel
Multiplex PCR panel for SARS-CoV-2 amplicon sequencing
The ARTIC panel consists of two primer pools that amplify SARS-CoV-2 viral genomes from complex samples for research. The sequencing data can elucidate the presence of mutations consistent with known variants or identify new variants in research studies.
xGen NGS—made for COVID-19 research.
xGen™ SARS-CoV-2 Midnight Amplicon Panel
Multiplex PCR primers for amplicon sequencing
Panel for amplification of SARS-CoV-2 from research samples that can be used to understand the spread of the virus, identify variants, and helps enable a better understanding of COVID-19.
xGen™ NGS—made for COVID-19 research.
xGen™ HS EGFR Pathway Amplicon Panel
Targeted sequencing panel for EGFR, BRAF, KRAS, and NRAS
xGen HS EGFR Pathway Amplicon Panel can identify variants with a MAF of ≤1% from ≥ 10 ng of cell-free DNA (cfDNA) or FFPE DNA for Illumina® sequencers.
xGen NGS—made sensitive.
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend for these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.
99.7% SARS-CoV-2 genomic coverage
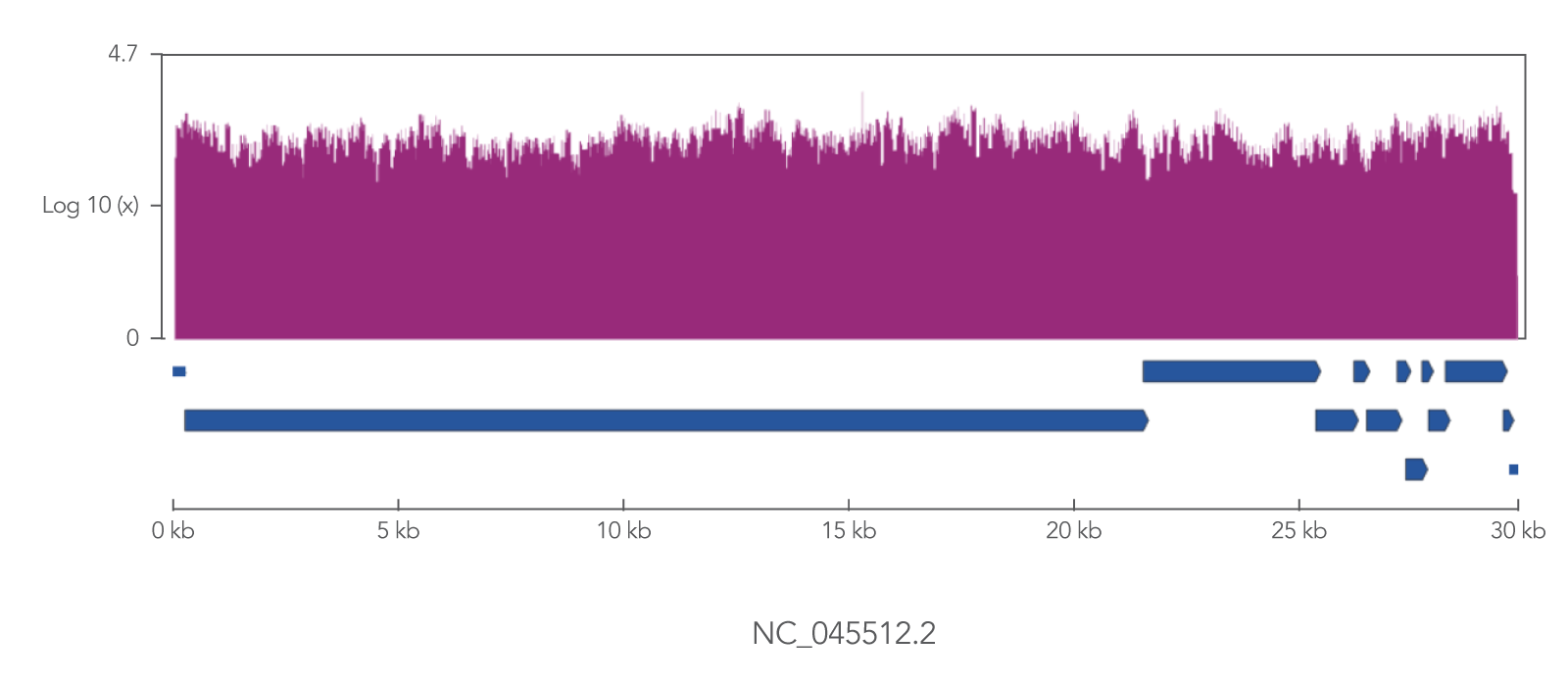
The xGen SARS-CoV-2 Amplicon Panel uses overlapping primers to generate 345 amplicons, sized 116–255 bp (average 150 bp), along the length of the 29.9 kb viral genome, obtaining 99.7% coverage of the genome (Figure 2). Overlapping primers ensure that variants are identified, even when the mutation interferes with a primer binding site. Comprehensive mutation identification is crucial for tracking nucleotide variants and improve understanding of virus evolution, transmission, and pathogenesis.
Figure 2. The xGen SARS-CoV-2 Amplicon Panel provides 99.7% coverage of the SARS-CoV-2 genome. RNA was isolated from gamma-irradiated and sonicated cell lysate from Vero E6 cultured monkey kidney epithelial cells (ATCC® CRL-1586™) infected with SARS-CoV-2 isolate USA-WA1/2020 (BEI Resources, Cat. No. NR-52287). 100,000 copies of SARS-CoV-2 RNA was converted into cDNA using the SuperScript™ IV Kit (Thermo Fisher Scientific). The resulting NGS library generated with the xGen SARS-CoV-2 Amplicon Panel was sequenced 2 x 150 bp on a MiniSeq® System (Illumina). Resulting reads were downsampled to 280k reads per sample (n = 2) for analysis. Genomic coverage for a representative sample is shown.
Full genomic coverage of lineages with a single primer set
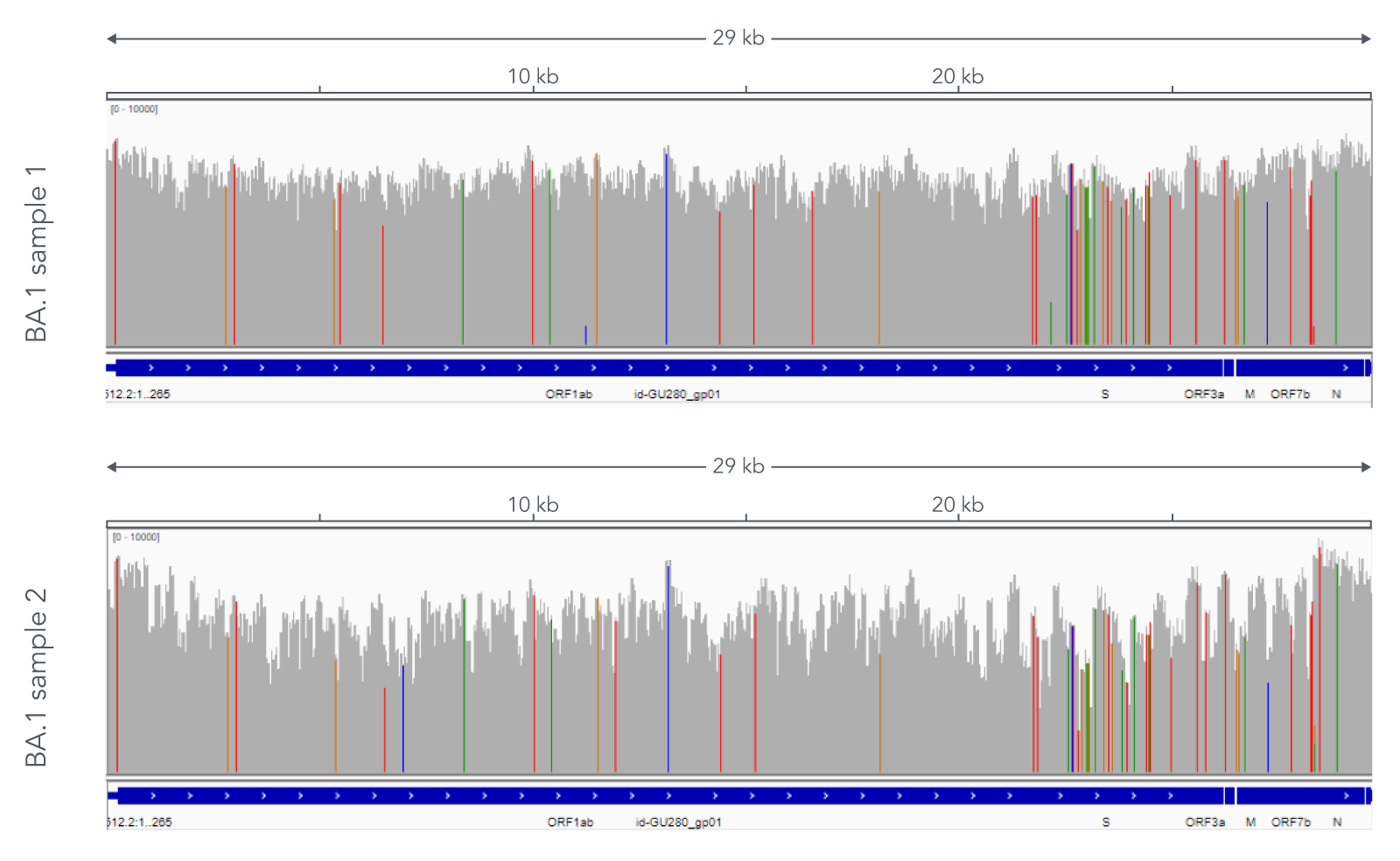
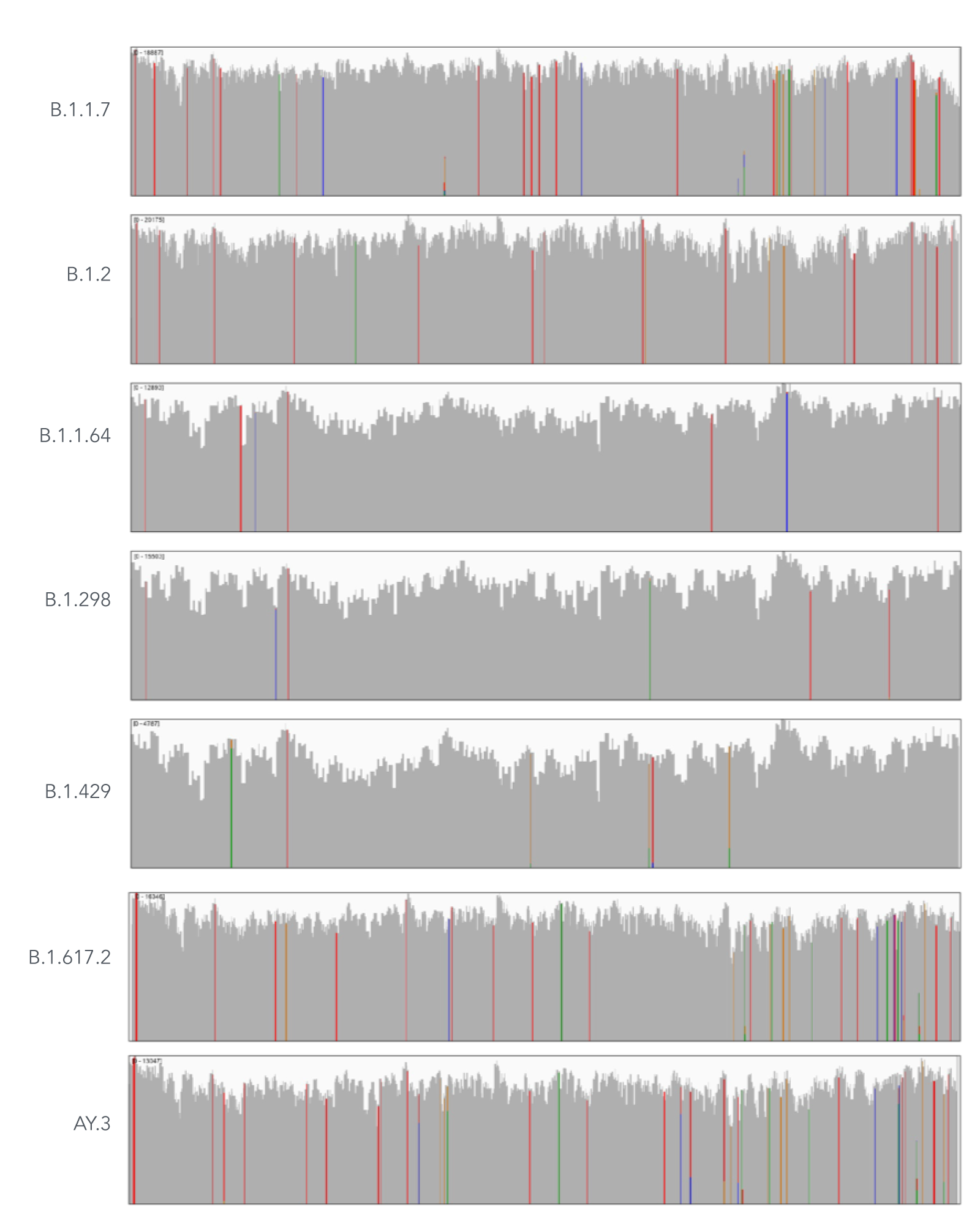
Using a single, consistent primer set, xGen SARS-CoV-2 Amplicon Panel has been shown to cover multiple lineages, including sub-lineages of Omicron (Figure 3), Delta, and Alpha (Figure 4). The primer set has remained unchanged since its release in 2020, preventing the need for frequent changes to primer designs.
Figure 3. Two representative examples of sequence coverage obtained from Omicron SARS-CoV-2 RNA. RNA was extracted from nasopharyngeal swabs and converted into cDNA using the SuperScript IV Kit (Thermo Fisher Scientific). The resulting library generated with the xGen SARS-CoV-2 Amplicon Panel was sequenced 2 x 150 bp on a MiniSeq System (Illumina). Resulting reads were downsampled to 83,000 reads per sample for analysis. Genomic coverage for two BA.1 lineages is shown. Ct values are 13.6 and 19.9. A total of 93 biological research samples that exhibited S gene dropout by qPCR were analyzed, 90 of which were identified as Omicron variants.
Figure 4. An example of coverages obtained from B.1.1.7 (Alpha), B.1.2, B.1.1.64, B.1.298, B.1.429 (Epsilon), B.1.617.2 (Delta), and AY.3 (Delta) SARS-CoV-2. RNA was extracted from nasopharyngeal or oropharyngeal swabs and converted into cDNA using the SuperScript IV Kit (Thermo Fisher Scientific). The resulting library generated with the xGen SARS-CoV-2 Amplicon Panel was sequenced 2 x 150 bp on a MiniSeq System (Illumina). Resulting reads were downsampled to 83,000 reads per sample for analysis. Genomic coverage for a representative sample per lineage is shown.
Research Investigation
Case Study I Genemarkers, LLC
Genemarkers, LLC, and Swift Biosciences collaborated to identify mutations and variants from SARS-CoV-2 samples sourced in Kalamazoo, Michigan. Samples were anticipated to be U.K. variants (B.1.1.7) due to the observation of S gene dropout in PCR testing. After generating an NGS library with the xGen SARS-CoV-2 Amplicon Panel and sequencing, the mutation pattern of SARS-CoV-2 B.1.1.7 was not observed in one sample, leading to clarification on variants present in the Kalamazoo area.
Obtain genomes from viral titers as low as 10–100 viral copies
10 to 1 million viral genome copies (Figure 5) are sufficient to generate NGS libraries using the xGen SARS-CoV-2 Amplicon Panel. Mixed RNA samples were converted into first strand cDNA and used to create sequencing libraries with the xGen SARS-CoV-2 Amplicon Panel. Libraries were enzymatically normalized to 4 nM using the Normalase workflow.
Figure 5. Obtain genomes from as few as 10–100 viral copies. SARS-CoV-2 RNA USA-WA1/2020 (BEI Resources, catalog # NR-52287) was mixed with Universal Human Reference RNA (Agilent, catalog # 740000) and converted into first-strand cDNA using the Superscript® IV First-Strand Synthesis System (Thermo Fisher Scientific). The resulting library generated with the xGen SARS-CoV-2 Amplicon Panel was sequenced on a MiniSeq System (Illumina) at 2 x 150 bp. Resulting reads were downsampled to 280k reads per sample (n = 2). Genomic copies identified are shown.
The recent emergence of monkeypox viral infections globally has resulted in an increased need for rapid, reliable NGS approaches to not only monitor and trace outbreaks but to also track any potentially novel variants that may arise. Epidemiological studies are currently underway to pinpoint transmission and infection patterns of this zoonotic disease. IDT recognizes the importance of these studies and has designed an xGen NGS Amplicon Sequencing Panel to target the monkeypox virus†.
The xGen Monkeypox Virus Amplicon Panel offers a streamlined (DNA-to-sequencer in 2.5 hours), single-tube NGS workflow for studying the monkeypox virus (MPXV). This Predesigned xGen Amplicon Panel provides 184 kb of high-quality coverage of the monkeypox genome (Table 2, Figure 2 and Figure 3) from inputs as low as 300 viral genome copies (Table 3). xGen Amplicon technology includes amplicon tiling and creation of super amplicons to ensure comprehensive genome coverage and provide resistance to future viral mutations that may fall on a priming site (Figure 4), thus enabling future identification of novel variants.
Figure 2. Coverage of the monkeypox virus genome. As an example of comprehensive coverage, at the specified positions, of monkeypox (DQ011157), ~3000 copies of the monkeypox genome (BEI Resources, NIAID, NIH: Genomic DNA from Monkeypox Virus, USA-2003, NR-4928) and 10 ng Coriell DNA NA12878 (human) were input into the xGen Amplicon workflow using the xGen Monkeypox Virus Amplicon Panel. The resulting NGS library was sequenced on a MiSeq system (Illumina) with 250 bp paired-end (PE) sequencing with 3,055,632 total reads. Reads were aligned and mapped to the monkeypox reference genome (DQ011157 [2]) using bwa (v 2.2.1 [3]). The total sequencing depth per genomic position (in log10 scale) was then plotted to visualize genomic coverage. Example data for one replicate are shown here, though experimental duplicates showed similar results.
Coverage and on-target mapping rates
Based on initial research and development, this panel has been shown to offer comprehensive coverage of the monkeypox viral genome from positions 6760–190,905. The inverted terminal repeats (ITRs) at both ends of the genome were omitted from the panel design due to the repetitive nature of these sequences.
To prepare amplicon sequencing libraries using the xGen Monkeypox Virus Amplicon Panel, ~3000 copies of the monkeypox genome (BEI Resources) and 10 ng Coriell DNA NA12878 (human) were used. The resulting NGS library was sequenced on a MiniSeq™ system (Illumina) with 150 bp paired-end (PE) sequencing with 1,774,058 total reads. Reads were aligned and mapped to the monkeypox reference genome (DQ011157 [2]) using bwa (v 2.2.1 [3]). Table 2 shows representative metrics obtained in this proof-of-concept experiment.
Table 2. xGen Monkeypox Virus Amplicon Panel NGS metrics.
| % mapping | % on-target (base) | % base uniformity (>0.2X mean) | |
|---|---|---|---|
| xGen Monkeypox Virus Amplicon Panel | 88.1 | 97.7 | 98.0 |
Figure 3. The xGen Monkeypox Virus Amplicon Panel results in comprehensive coverage of the monkeypox genome (excluding the ITRs). To prepare amplicon sequencing libraries using the xGen Monkeypox Virus Amplicon Panel, ~3000 copies of the monkeypox genome (BEI Resources, NIAID, NIH: Genomic DNA from Monkeypox Virus, USA-2003, NR-4928) and 10 ng Coriell DNA NA12878 (human) were used. The resulting NGS library was sequenced on a MiniSeq system (Illumina) with 150 bp paired-end (PE) sequencing with 1,774,058 total reads. Reads were aligned and mapped to the monkeypox reference genome (DQ011157 [2]) using bwa (v 2.2.1 [3]). The shown dot plot indicates the relative coverage of each amplicon in the panel (n = 4), where each amplicon is represented by a blue dot. Colored dashed lines represent mean coverage of 0.05X, 0.1X, 0.2X, 1X, and 5X.
Sequencing results from titers as low as 300 viral genome copies
The input into xGen Monkeypox Virus Amplicon Panel consisted of either 3000, 300, or 0 copies of the monkeypox genome (BEI Resources) and 10 ng Coriell DNA NA12878 (human). The resulting NGS library was sequenced as described above. Reads were aligned and mapped to the monkeypox reference genome (DQ011157 [2]) using bwa (v 2.2.1 [3]). A high level of genomic coverage was observed with 3000 and 300 monkeypox genomic copies (Table 3), and no genomic coverage was observed with no monkeypox copy input.
Table 3. The xGen Monkeypox Virus Amplicon Panel provides coverage for a range of viral DNA inputs.
| Sample number | Input copies of viral genomes | Total reads |
Percent target bases 10X (base) |
|---|---|---|---|
| 1 | 3000 | 2,374,500 | 99.4 |
| 2 | 2,725,540 | 99.4 | |
| 3 | 300 | 1,984,616 | 98.8 |
| 4 | 1,963,414 | 98.7 | |
| 5 | 0/NTC | 2,183,412 | 0.2 |
| 6 | 2,464,906 | 0.3 |
Super amplicons help maintain sequencing coverage despite mutations
Figure 4. The xGen Monkeypox Virus Amplicon Panel maintains genomic coverage despite mutations at primer binding sites. The generation of super amplicons means that even in the case of a mutation occurring in primer binding sites (shown here by black arrows), the xGen Monkeypox Virus Amplicon Panel can maintain genomic coverage. The input into library prep consisted of ~3000 copies of the monkeypox genome (BEI Resources) and 10 ng Coriell DNA NA12878 (human). The resulting NGS library was sequenced on a MiniSeq system (Illumina) (150 bp PE sequencing) with 1,774,058 total reads. Reads were aligned and mapped to the monkeypox reference genome (DQ011157 [2]) using bwa (v 2.2.1 [3]) and coverage was visualized with IGV (Broad Institute [4]).
| Features | xGen 16S v2 and ITS1 Amplicon Panels |
|---|---|
| Panel information | 23 primers (16S v2); average 425 bp amplicon size 15 primers (ITS1); amplicon size 145–695 bp |
| Input material | 10 pg for microbial isolates; 1–50 ng for metagenomic samples |
| Time | 2 hours cDNA-to-library or 3 hours cDNA-to-normalized-library-pool |
| Components available |
Note: Kits do not include RT module or magnetic beads |
| Multiplexing capability | Up to 96 combinatorial dual index (CDI) or 1536 unique dual index (UDI) |
| Recommended depth | 16S v2: 100K reads per library ITS1: 25K reads per library |
Coverage of all variable regions of the 16S rRNA gene and ITS1 region
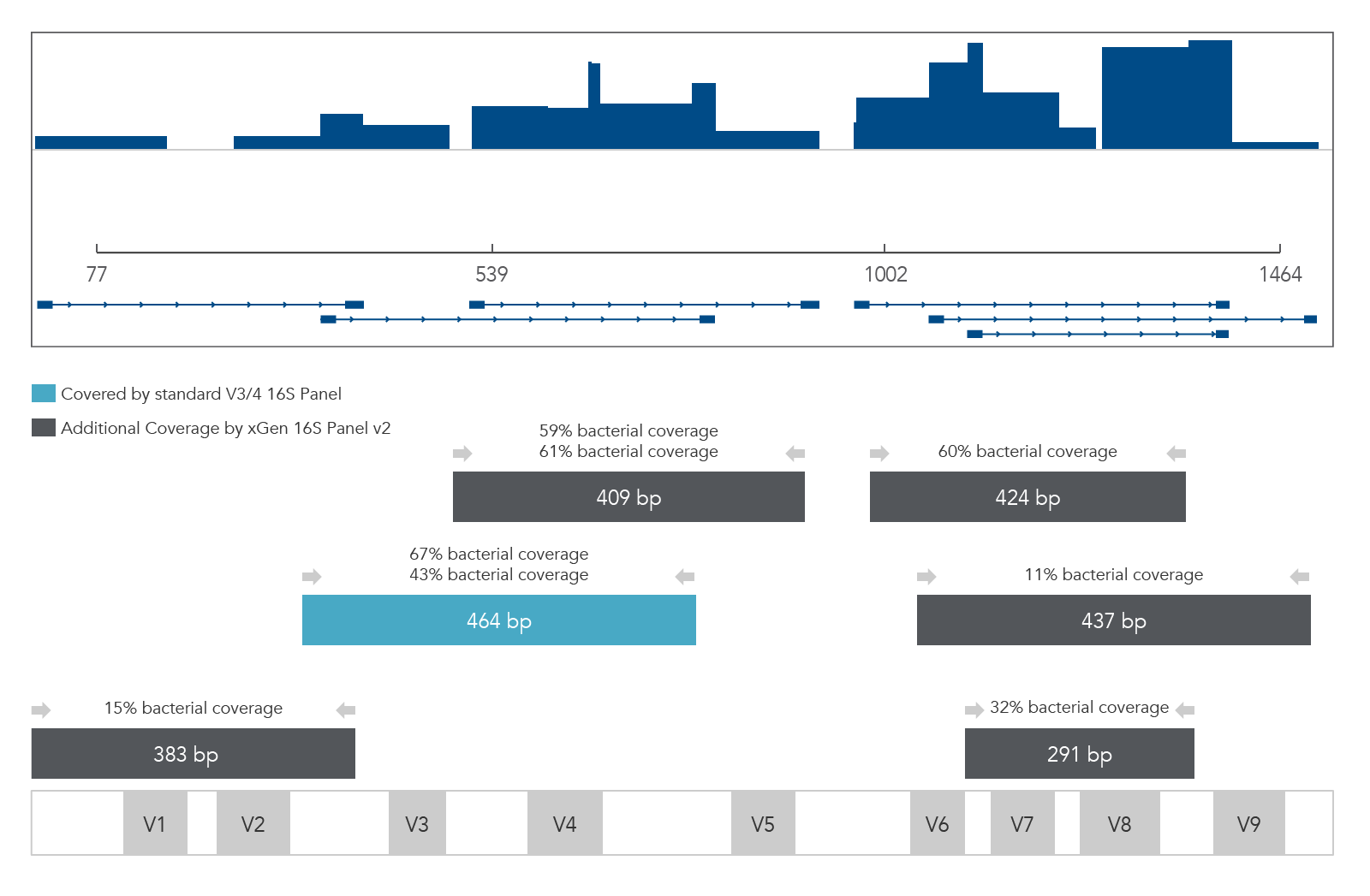
The 16S v2 and xGen ITS1 Amplicon Panels facilitate NGS analysis of complex microbial communities (e.g., bacteria, archaea, fungi) using primer pools that target the 16S rRNA gene (variable regions 1–9) and ITS1 region (Figure 2). In addition, these panels can be customized with additional targets, including antibiotic resistance or virulence genes, allowing sub-genera level identification and functional analysis.
xGen 16S Amplicon Panel v2 (primers)
Figure 2. xGen 16S Amplicon Panel v2 (primers). Sequencing read coverage for an E. coli DNA sample (n = 1) observed in Integrative Genomics Viewer (IGV) Sashimi plot and illustration of multiplexed primer coverage of all nine variable regions of 16S rRNA compared to a standard V3/V4 16S sequencing read coverage.
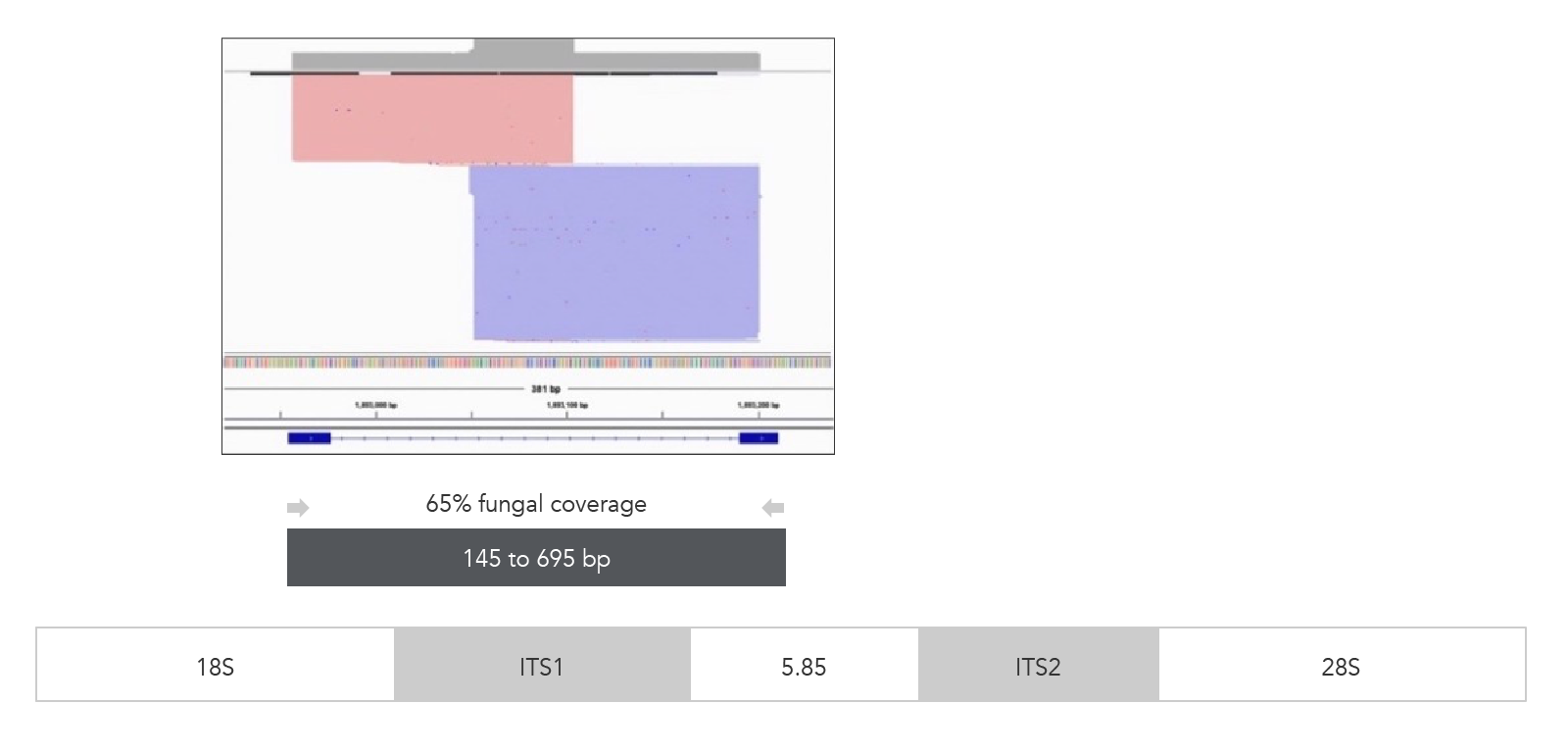
xGen ITS1 Amplicon Panel (primers)
Figure 3. xGen ITS1 Amplicon Panel (primers). Sequencing read coverage observed in the IGV Sashimi plot and illustration of multiplexed primer coverage of ITS1 region for Candida albicans (n = 1). Reads originating from the forward primer are shown in red; reads from the reverse primer are in blue.
The xGen 16S Amplicon Panel v2 and the xGen ITS1 Amplicon Panel provide representation of diverse microbial communities
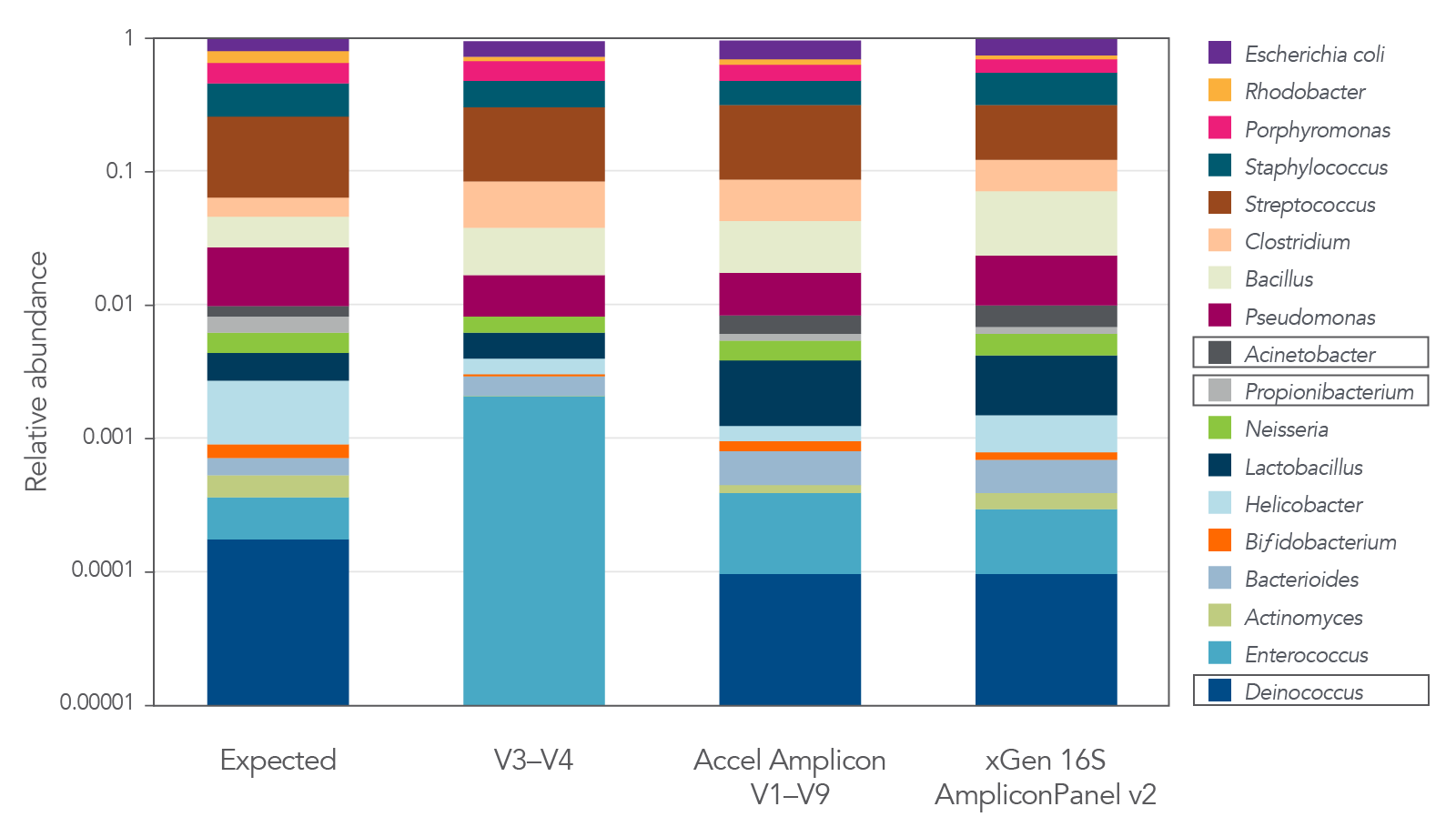
Figure 4. The xGen 16S Amplicon Panel v2 provides superior representation of diverse microbial communities. The 16S v2 panel covering V1–V9 regions of 16S rRNA provides accurate representation of each genus in a commercially available standard (MSA-1003) compared to libraries interrogating the V3–V4 region alone. The legacy Accel-Amplicon 16S+ITS panel and the new xGen 16S Amplicon Panel v2 show similar relative abundance results. Strains were present at levels from 0.02% to 18% in MSA-1003. Boxed organisms were not detected with sole use of the V3-V4 region. Libraries were sequenced on a MiSeq® (Illumina) instrument without the use of the PhiX reagent. As shown in the examples in this figure, by not losing reads to PhiX or having to sequence deeper due to the use of phased primers, an increased number of samples can be fit on a sequencing run, thereby increasing sequencing efficiency, while still achieving quality data.
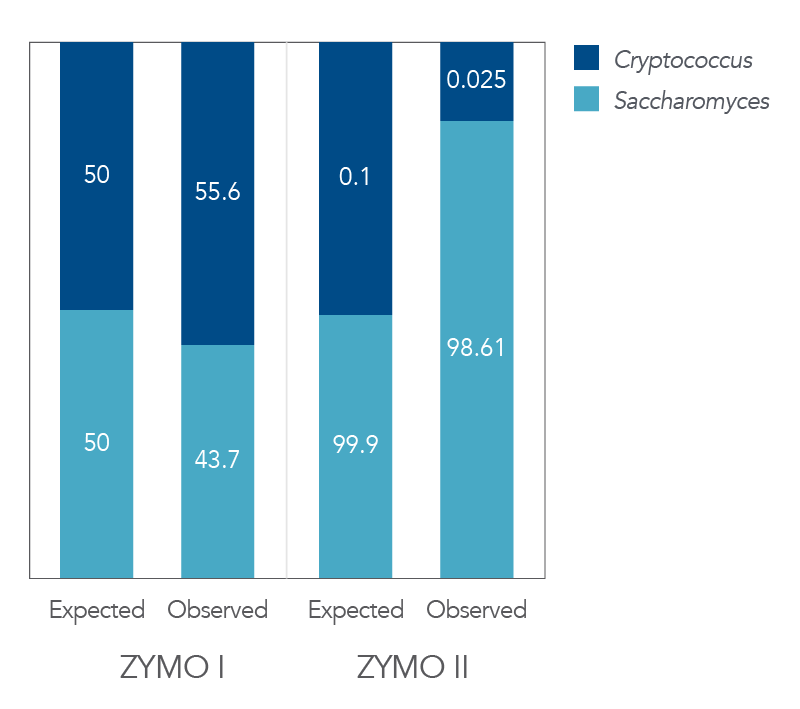
Figure 5. Expected vs. observed Zymo I/II data from the panel covering the ITS1 region. The xGen ITS1 Amplicon Panel provides accurate representation of each genus in two commercially available standards [Zymo I—ZymoBIOMICS™ Microbial Community standard and Zymo II—ZymoBIOMICS Microbial Community standard II (Log distribution)]. Fungal strains were present at levels from 0.1% to 99.9% in Zymo II. The xGen ITS1 Amplicon Panel resulted in expected representation of the fungal species in a bacterial background.
Consistent function with varying biomass, sample types, and read length
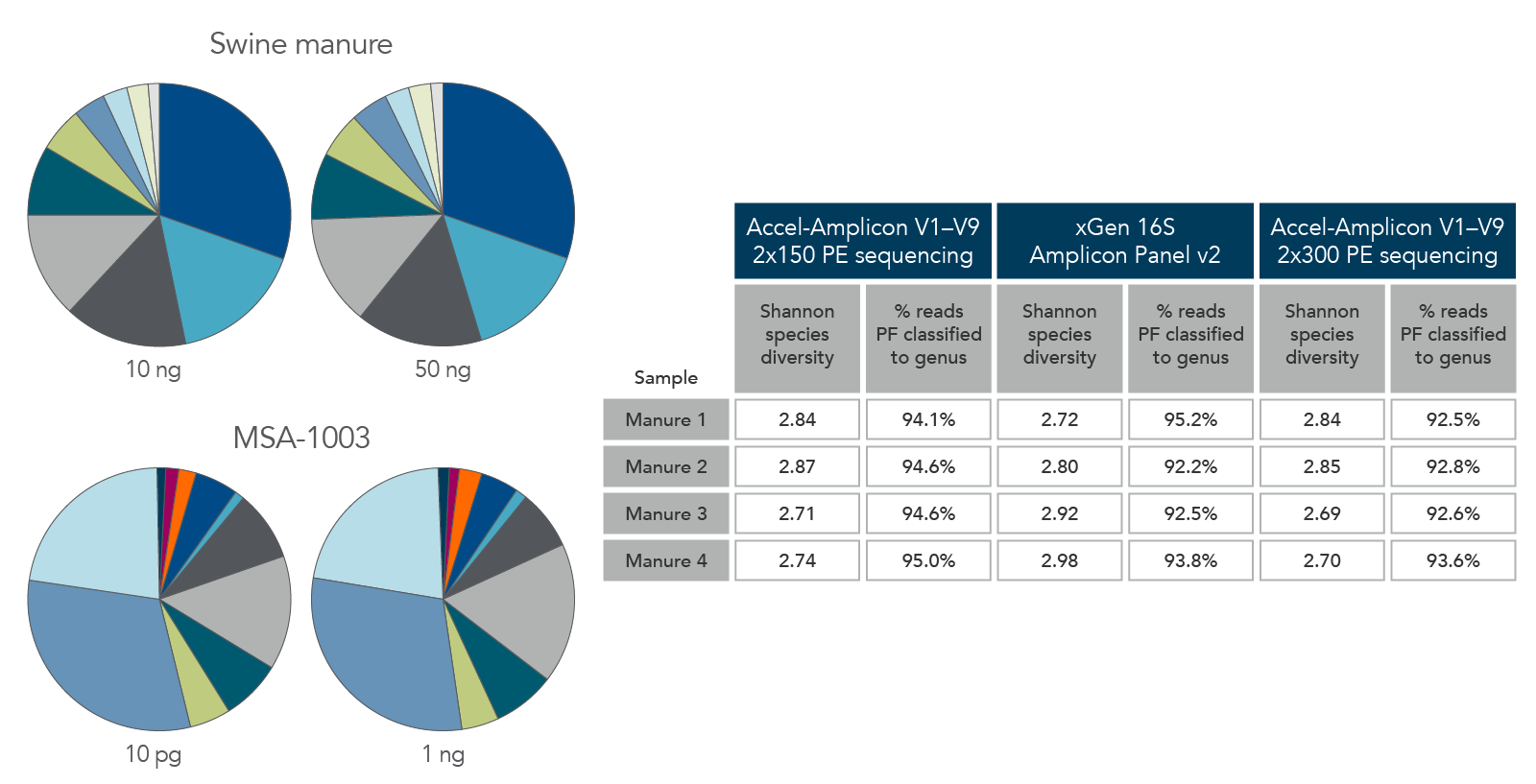
Figure 6. Consistent function with varying biomass, sample type, and read length. Using the same protocol and cycling conditions, input quantities of 10 ng to 50 ng of swine manure (top left) identified the same quantities and types of genera. In addition, the 10 pg sample of MSA-1003 (bottom left) had the same relative abundance and type of genera as the 1 ng sample. The legacy Accel-Amplicon 16S+ITS panel and the new xGen 16S Amplicon Panel v2 products gave similar results when comparing the 2 x 150 and 2 x 300 PE sequencing read percentages; a comparable number of genera were identified from swine manure samples (right table).
Table 1. Reliable results across variety of population groups and sex.
| DNA | Sex | Race/population group | % On target | % Coverage uniformity |
|---|---|---|---|---|
| NA00897 | M | Caucasian | 99.7 | 98.2 |
| NA11496 | F | Caucasian | 96.2 | 100 |
| NA14624 | M | Caucasian | 99.9 | 93.3 |
| NA24143 | F | Caucasian (Ashkenazi) | 99.8 | 93.4 |
| NA14639 | F | Caucasian (Ashkenazi) | 99.9 | 93.3 |
| NA14628 | F | Caucasian (English/German) | 99.9 | 93.3 |
| NA14637 | F | Caucasian (French Canadian) | 99.9 | 94.5 |
| NA14634 | M | Caucasian (Norwegian) | 99.9 | 95.6 |
| NA12878 | F | Caucasian (Utah/Mormon) | 99.8 | 99.3 |
| NA12249 | F | Caucasian (Utah/Mormon) | 99.9 | 93.3 |
| HG02190 | F | Chinese (Dai) | 99.9 | 93.3 |
| NA24695 | F | Chinese (Han) | 99.9 | 92.2 |
| NA19240 | F | Yoruban | 99.7 | 97.1 |
Sequencing metrics for the xGen Sample ID Amplicon Panel with an array of Coriell repository DNA samples (10 ng input each) of different population groups and sex. Libraries were sequenced on a MiniSeq® (Illumina) instrument. On-target and coverage uniformity were calculated as per base.
| Chr | POS | SNP ID | REF | ALT | Coriell NA1278 | Coriell NA00897 | Coriell NA11496 |
|---|---|---|---|---|---|---|---|
| 1 | 67861520 | rs2229546 | C | A | 100% | 52% | |
| 1 | 158582646 | rs2251969 | T | C | 100% | ||
| 1 | 167849414 | rs203849 | A | G | 50% | 100% | |
| 1 | 179520506 | rs1410592 | G | A | 100% | 53% | 100% |
| 1 | 209811886 | rs2076356 | T | G | 53% | 52% | |
| 1 | 209968684 | rs2013162 | C | A | 100% | 47% | |
| 1 | 228431095 | rs1771455 | A | G | 47% | 100% | 53% |
| 2 | 44502788 | rs3738985 | A | C | 100% | 100% | 100% |
| 2 | 49381585 | rs1394205 | C | T | 47% | ||
| 2 | 75115108 | rs10194657 | A | G | 50% | ||
| 2 | 169789016 | rs497692 | T | C | 100% | ||
| 2 | 170092395 | rs2229267 | A | G | 52% | 47% | 51% |
| 2 | 179454394 | rs1560221 | A | G | 47% | 48% | |
| 2 | 179455207 | rs2163009 | T | C | 45% | 48% | |
| 2 | 215820013 | rs10498027 | G | A | 45% | ||
| 2 | 219941063 | rs897477 | G | A | 100% | 100% | 100% |
| 2 | 227896976 | rs10203363 | C | T | 100% | 46% | |
| 3 | 4403767 | rs2819561 | A | G | 50% | 100% | 51% |
| 3 | 4712413 | rs2306875 | G | A | 48% | 52% | 45% |
| 3 | 45989044 | rs2234358 | T | G | 49% | 100% | |
| 3 | 148727133 | rs4938 | G | A | 52% | 51% | 46% |
| 4 | 5749904 | rs386594666 | T | C | 50% | 49% | |
| 4 | 86844835 | rs6824722 | A | G | 47% | ||
| 4 | 86915848 | rs10003909 | T | C | 51% | 50% | |
| 4 | 88534235 | rs2736982 | A | G | 100% | 100% | |
| 5 | 13719022 | rs30169 | T | G | 100% | 100% | 48% |
| 5 | 13829799 | rs1348689 | G | A | 50% | 50% | 100% |
| 5 | 55155402 | rs1009639 | C | T | 99% | 99% | 99% |
| 5 | 82834630 | rs309557 | T | C | 53% | 54% | |
| 5 | 135392426 | rs4669 | T | C | 48% | ||
| 5 | 138456815 | rs3088052 | T | C | 48% | 52% | 52% |
| 6 | 146755140 | rs2942 | G | A | 46% | 47% | 49% |
| 6 | 152464839 | rs2256135 | A | G | 51% | 52% | 49% |
| 7 | 34009946 | rs10265207 | C | T | 100% | 99% | |
| 7 | 48450157 | rs17548783 | T | C | 47% | 100% | |
| 7 | 50742180 | rs1800504 | C | T | 50% | 50% | 100% |
| 7 | 100804140 | rs1048303 | C | T | 52% | 100% | |
| 7 | 127250907 | rs386607686 | T | C | 46% | 100% | 100% |
| 8 | 94935937 | rs4735258 | T | C | 100% | 51% | |
| 8 | 104337096 | rs3808554 | A | G | 48% | 47% | 45% |
| 9 | 27202870 | rs386602523 | A | G | 50% | 46% | |
| 9 | 77415284 | rs7859201 | A | C | 100% | ||
| 9 | 97365642 | rs9695 | G | A | 47% | 51% | |
| 9 | 100190780 | rs1381532 | A | G | 100% | 100% | |
| 9 | 104184022 | rs4577 | G | A | 100% | ||
| 9 | 136304497 | rs3124768 | A | G | 100% | 47% | 52% |
| 10 | 69926097 | rs2673794 | T | C | 47% | 100% | |
| 10 | 73856984 | rs3312 | A | G | 45% | ||
| 10 | 78944590 | rs1131824 | G | A | 47% | 49% | 100% |
| 10 | 85972043 | rs10749482 | A | G | 51% | 47% | 46% |
| 10 | 95791763 | rs17109674 | G | A | 50% | ||
| 10 | 104596924 | rs6163 | C | A | 47% | ||
| 10 | 104814162 | rs2275271 | T | C | 46% | ||
| 10 | 105819956 | rs805701 | G | A | 100% | 100% | 100% |
| 10 | 113920465 | rs2277207 | G | A | 48% | 47% | 47% |
| 11 | 6629665 | rs1043388 | C | T | 47% | ||
| 11 | 16133413 | rs4617548 | A | G | 100% | 45% | |
| 11 | 30255185 | rs386601843 | C | T | 51% | 48% | 100% |
| 12 | 993930 | rs7300444 | C | T | |||
| 12 | 8757481 | rs2028373 | G | A | 53% | 41% | |
| 12 | 52200742 | rs60637 | C | A | 50% | 100% | 100% |
| 14 | 50769717 | rs2297995 | G | A | 100% | 100% | |
| 14 | 64637147 | rs7161192 | C | A | 47% | ||
| 14 | 74992800 | rs699374 | A | G | 51% | 45% | |
| 14 | 76045858 | rs2287016 | G | A | |||
| 15 | 34528948 | rs4577050 | G | A | 100% | 47% | 46% |
| 15 | 89401615 | rs3825994 | T | G | 100% | 48% | 100% |
| 15 | 89402596 | rs698621 | T | G | 53% | 48% | 100% |
| 16 | 68713730 | rs2296409 | G | A | 51% | 100% | 100% |
| 16 | 68713823 | rs2296408 | C | A | 49% | 100% | 100% |
| 16 | 68729785 | rs17715450 | C | A | 48% | 100% | 100% |
| 16 | 70303580 | rs2070203 | G | A | 47% | 48% | 46% |
| 16 | 70546234 | rs3762171 | G | A | 48% | 45% | 50% |
| 17 | 7192091 | rs222842 | C | T | 47% | 100% | 100% |
| 17 | 10536018 | rs2285479 | G | A | 100% | 100% | |
| 17 | 10542471 | rs2285475 | T | G | 100% | 100% | |
| 17 | 42449789 | rs5910 | G | A | 100% | 48% | |
| 17 | 71192663 | rs1052706 | G | A | 100% | 51% | 55% |
| 17 | 71192873 | rs11544800 | A | G | 100% | 53% | 52% |
| 17 | 71197748 | rs1037256 | G | A | 100% | 49% | 56% |
| 18 | 12351342 | rs11080572 | C | T | 47% | 100% | 52% |
| 18 | 47455923 | rs2298628 | C | T | 48% | 52% | 100% |
| 19 | 10267077 | rs2228611 | T | C | 48% | 52% | 100% |
| 19 | 12989560 | rs2293682 | G | A | 47% | 51% | |
| 19 | 13445208 | rs2248069 | C | T | 100% | 100% | 46% |
| 19 | 16591464 | rs9305079 | G | A | 100% | 50% | 49% |
| 19 | 38994910 | rs2229144 | G | A | 51% | ||
| 20 | 2413320 | rs2076652 | T | C | 52% | 100% | 51% |
| 20 | 6100088 | rs10373 | A | G | 51% | 45% | 47% |
| 20 | 19970705 | rs2076584 | C | T | 100% | 100% | 51% |
| 20 | 35865054 | rs4608 | C | T | 100% | 48% | 100% |
| 21 | 44323590 | rs4148973 | T | G | 48% | 51% | 48% |
| 21 | 46908355 | rs11702425 | T | C | 51% | 51% | 47% |
| 21 | 47773103 | rs2249057 | C | A | 100% | 47% | 50% |
| 22 | 21141300 | rs4675 | T | C | 51% | 51% | 48% |
| 22 | 37469591 | rs4820268 | G | A | 51% | 100% | 50% |
The xGen Sample ID Amplicon Panel was used with 10 ng of gDNA from different Coriell samples to create libraries. Sequencing was performed using MiSeq® V2 Reagents. SNP allele frequencies were determined using GATK HaplotypeCaller (Broad Institute). Homozygous SNPs are indicated in blue and heterozygous SNPs are indicated in green. Positions on GHCh37 (hg19). Clear differences are observed between the DNA samples, demonstrating the genotype determination possible with xGen Sample ID Amplicon Panel.
Amplicon distribution per chromosome
Amplicons per chromosome in xGen Sample ID Amplicon Panel consists of 104 amplicons, 95 of which cover known exonic SNPs, and 9 are for sex identification. This coverage gives a discrimination power of 1:85,000 [1].
| Chr | #amps |
|---|---|
| X | 1 |
| Y | 8 |
| 1 | 8 |
| 2 | 21 |
| 3 | 5 |
| 4 | 5 |
| 5 | 7 |
| 6 | 3 |
| 7 | 6 |
| 8 | 3 |
| 9 | 7 |
| 10 | 9 |
| 11 | 4 |
| 12 | 4 |
| 13 | 1 |
| 14 | 5 |
| 15 | 4 |
| 16 | 6 |
| 17 | 8 |
| 18 | 3 |
| 19 | 5 |
| 20 | 5 |
| 21 | 4 |
| 22 | 3 |
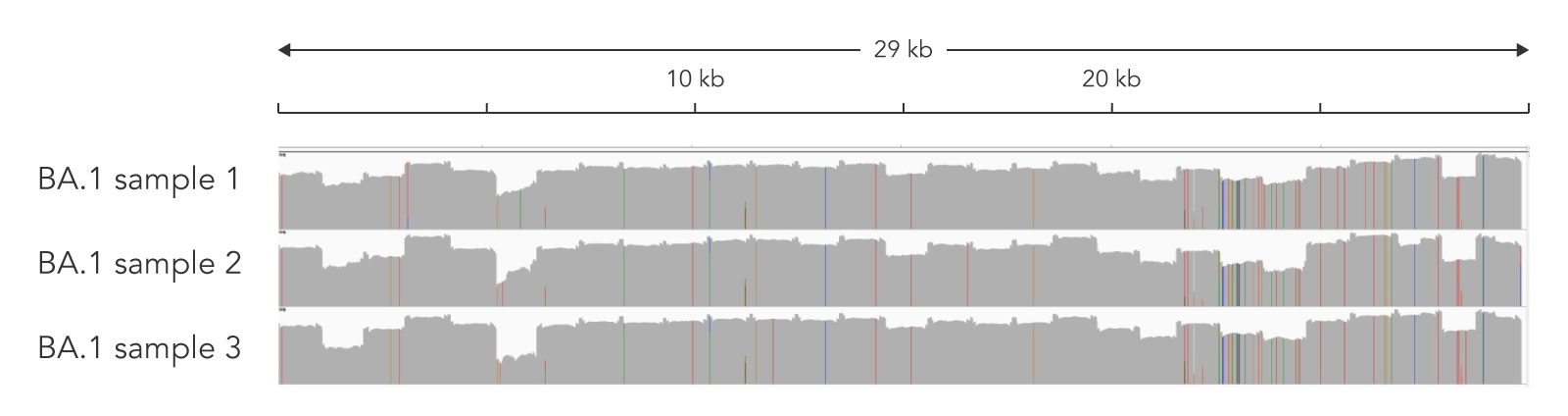
Figure 1. The ARTIC SARS-CoV-2 Amplicon Panel’s genome coverage for Omicron variants. Two nasopharyngeal swab samples (Cts 28.4 and 13.6) were subjected to the protocol using the ARTIC v4.1 primer pool to create amplicons, then the xGen DNA Library Prep MC Library Prep Kit to adapt the amplicons to create an NGS library. Libraries were sequenced on a MiSeq™ (Illumina) with 2 x 250 bp PE reads. Data was down sampled to 100,000 reads. Alignment was then performed using the Burrows-Wheeler Alignment (BWA) Tool [4] and a consensus file uploaded to Pangolin for lineage calling. High genomic coverage for the BA.1 lineage was observed (IGV screenshot above) indicating the compatibility of the panel with Omicron variants.
Table 2. Sequencing metrics for ARTIC v4.1 libraries created from nasopharyngeal swabs.
| Sample | Ct | Reads aligned | On-target | Coverage uniformity | % Genome >10X | Lineage |
|---|---|---|---|---|---|---|
| Sample 1 | 28.4 | 100,000 | 99.7 | 85.9 | 97 | BA.1 |
| Sample 2 | 13.6 | 100,000 | 99.7 | 97.6 | 100 | BA.1 |
Sequencing metrics for NGS libraries originating from nasopharyngeal swabs. Libraries were created with the ARTIC v4.1 primer pool and the xGen DNA Library Prep MC Library Prep Kit. Libraries were sequenced on a MiSeq™ (Illumina) with 2 x 250 bp PE reads. Data was down sampled to 100,000 reads. Lineages were called with Pangolin.
As previously noted, the Ct value for the viral RNA in the sample is a critical metric to obtain usable data. IDT researchers used the SARS-CoV-2 Midnight Amplicon Panel in combination with the IDT xGen DNA Library Prep Kit EZ on samples obtained from nasopharyngeal swabs.
In a proof-of-concept test, sequencing libraries were first prepared according to the SARS-CoV-2 Midnight Amplicon Panel protocol for samples with varying Ct values. These libraries were then subjected to enzymatic fragmentation and library preparation according to the xGen DNA Library Prep Kit EZ recommendations (Figure 3).
Figure 3. Amplicon sequencing combining the SARS-CoV-2 Midnight Amplicon Panel and the xGen DNA Library Prep Kit EZ provided >99.5% coverage at >10X of the SARS-CoV-2 genome for Ct values 14−19. Data was generated from three independent samples of nasopharyngeal (NP) swab material with three different Ct values: 14.1, 19.9, and 15.3. Libraries were generated following the xGen DNA Library Prep Kit EZ recommendations. The resulting libraries were sequenced with MiSeq™ 300 cycle kit (2 x 150 reads) (Illumina). Samples were subsampled to 197,000 total reads. 100.0% of the reads were aligned to the viral genome and the coverage uniformity ranged from 81−91.9% with a mean coverage of ~950X. Pangolin analysis indicated all samples were Omicron strain (BA.1).
To evaluate the xGen HS EGFR Pathway Amplicon Panel, two DNA standards from Seracare were used. These standards contain mutations with known allele frequencies at either 0.5% or 0.25%. Duplicate SeraSeq® ctDNA Reference Material AF (LGC SeraCare) samples at 10 ng were generated with the 0.5% standard and duplicate SeraSeq® ctDNA Reference Material AF (LGC SeraCare) samples at 20 ng were generated with the 0.25% standard, a total of 4 libraries are shown. As seen in Table 1, the observed allele frequency (AF) aligned with the expected allele frequency for the mutations in the reference cfDNA that were targeted by the xGen HS EGFR Pathway Amplicon Panel. Samples were sequenced on a MiniSeqTM (Illumina).
Table 1: Observed vs. expected allele frequency in NGS libraries prepared using the xGen HS EGFR Pathway Amplicon Panel
| 10 ng SeraSeq cfDNA with AF of 0.5% | 20 ng SeraSeq cfDNA with AF of 0.25% | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Mutation | Chr | Position | Expected AF | Observed AF (Rep 1) | Observed AF (Rep 2) | Expected AF | Observed AF (Rep 1) | Observed AF (Rep 2) |
| BRAF | p.V600E | 7 | 140453136 | 0.5% | 0.19% | 0.45% | 0.25% | 0.18% | 0.18% |
| EGFR | p.T790M | 7 | 55249071 | 0.5% | 0.43% | 0.57% | 0.25% | 0.33% | 0.36% |
| EGFR | p.L858R | 7 | 55259515 | 0.5% | 0.86% | 0.43% | 0.25% | 0.26% | 0.25% |
| KRAS | p.G12D | 12 | 25398284 | 0.5% | 0.59% | 0.47% | 0.25% | 0.20% | 0.48% |
| NRAS | p.Q61R | 1 | 115256529 | 0.5% | 0.91% | 0.56% | 0.25% | 0.30% | ND |
| EGFR | p.E746-A750 (delELREA) | 7 | 55242265 | 0.5% | 0.24% | 0.61% | 0.25% | 0.11% | 0.07% |
| EGFR | p.D770-N771 (ins) | 7 | 55249012 | 0.5% | 0.43% | 0.57% | 0.25% | 0.32% | 0.36% |
xGen SARS-CoV-2 Amplicon Panels, previously known as Swift Normalase™ Amplicon Panel (SNAP) SARS-CoV-2 enables researchers to identify SARS-CoV-2 strains, including variants, by creating next generation sequencing (NGS) libraries. The sequencing data can then answer research questions, such as:
- Which viral strains account for most infections within a local population?
- What new strains are emerging?
IDT’s predesigned amplicon panels for COVID research can aid in identifying the extent and pattern of viral spread within a population and allow researchers to quantify the effectiveness of intervention measures.
The xGen SARS-CoV-2 Amplicon Panel enables identification of SARS-CoV-2 variants from a wide array of research sample types such as nasopharyngeal/oropharyngeal swabs, sputa, stool, and wastewater. This panel covers 99.7% of the SARS-CoV-2 genome for identification of mutations from Alpha and Delta strains (see Product Data).
The xGen SARS-CoV-2 Sgene Amplicon Panel provides 100% coverage of the gene which encodes for the spike protein in the virus that causes COVID-19. Identify the presence of emerging viral strains which contain mutations in the Sgene and model potential changes in transmission patterns based upon those mutations.
The xGen ACE2 Gene Amplicon Panel contains 41 amplicons with an average size of 150 bp that provides comprehensive coverage of all coding regions of ACE2. Sequencing ACE2 has the potential to provide insight into disease outcome and facilitate further epidemiological investigations.
cDNA-to-sequencer in 3 hours
To sequence SARS-CoV-2 viral genomic material, first prepare first- or second-strand cDNA from samples such as nasopharyngeal/oropharyngeal swabs, sputa, stool, or wastewater (Figure 1). Generate an NGS library using tiled primer pairs in a single tube to target the 29.9 kb viral genome. Primers were designed against the NCBI Reference Sequence NC_045512.2 (severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome) (Table 1). Following a positive result by qPCR, use excess cDNA to perform variant calling and identify the viral strain.
Table 1. NGS for SARS-CoV-2 specifications.
| Features | Specifications |
|---|---|
| Design coverage and panel information |
|
| Input material |
|
| Time | 2 hours cDNA-to-Library |
| 3 hours cDNA-to-Normalized-Library Pool | |
| Multiplexing capability |
|
| Compatible with other indexes? | Yes |
| Recommended depth |
|
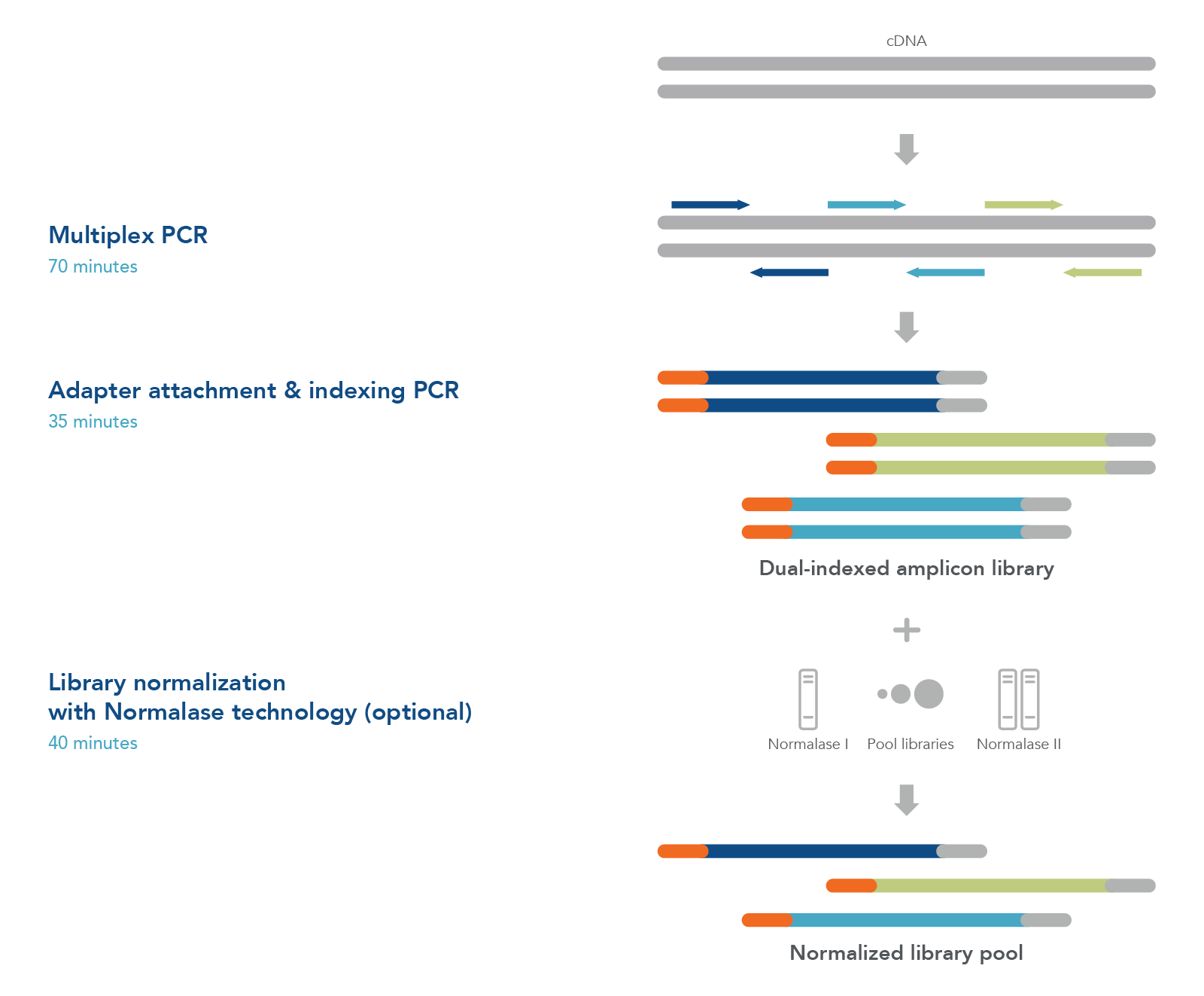
xGen Amplicon Panels for oncology and inherited diseases research utilize multiple overlapping amplicons in a single tube, using a rapid, 2-hour workflow to prepare ready-to-sequence libraries in research studies (Figure 1). The PCR1+PCR2 workflow generates NGS libraries for identifying genetic changes in the genes associated with a variety of tissues and inherited diseases (Table 1). The libraries may be quantified with conventional methods such as Qubit® (Thermo Fisher Scientific) or Agilent Bioanalyzer® and normalized by manual pooling or normalized enzymatically with the included xGen Normalase™ reagents.
Figure 1. xGen Amplicon Panels have a single tube workflow that is done in as little as 2 hours. Creating an NGS library starts with multiplex PCR. Your custom or predesigned panel is combined with the DNA sample to amplify the targets of interest. The samples are then amplified with indexing primers to create a functional dual indexed library. As an optional step, the xGen Normalase reagent can be used after pooling multiple libraries to ensure each is equally represented in the final sample for the flowcell.
Table 1. List of xGen Oncology & Inherited Diseases Amplicon Panels
| Panel | Number of Genes | Genes |
|---|---|---|
| xGen 56G Oncology Amplicon Panel v2, 96 rxn | 56 | ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CSF1R, CTNNB1, DDR2, DNMT3A, EGFR, ERBB2, ERBB4, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, FOXL2, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, IDH2, JAK2, JAK3, KDR, KIT, KRAS, MAP2K1, MET, MLH1, MPL, MSH6, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53, TSC1, VHL |
| xGen 57G Pan-Cancer Amplicon Panel, 96 rxn | 57 | ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CSF1R, CTNNB1, DDR2, DNMT3A, EGFR, ERBB2, ERBB4, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, FOXL2, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, IDH2, JAK2, JAK3, KDR, KIT, KRAS, MAP2K1, MET, MLH1, MPL, MSH6, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53, TSC1, TSC2, VHL |
| xGen BRCA1 BRCA2 Amplicon Panel, 96 rxn | 2 | BRCA1, BRCA2 |
| xGen BRCA1 BRCA2 PALB2 Amplicon Panel, 96 rxn | 3 | BRCA1, BRCA2, PALB2 |
| xGen CFTR Amplicon Panel, 96 rxn | 1 | CFTR (All exons including 5’ and 3’ UTRs, select intronic regions (1, 12, 22, and 25)) |
| xGen EGFR Pathway Amplicon Panel, 96 rxn | 4 | EGFR, BRAF, KRAS, NRAS |
| xGen Colorectal Amplicon Panel, 96 rxn | 16 | AKT1, APC, BRAF, ERBB2, ERBB4, KIT, KRAS, NOTCH1, NRAS, PDGFRA, PIK3CA, POLE, PTEN, SMAD4, STK11, TP53 |
| xGen TP53 Amplicon Panel, 96 rxn | 1 | TP53 |
| xGen Lung Amplicon Panel, 96 rxn | 17 | AKT1, ALK, ARAF, BRAF, EGFR, ERBB2, ERBB4, FGFR1, FGFR2, FGFR3, KRAS, MAP2K1, MET, NRAS, PIK3CA, PTEN, TP53 |
| xGen Lynch Syndrome Amplicon Panel, 96 rxn | 4 | MLH1, MSH2, MSH6, PMS2 |
| xGen Myeloid Amplicon Panel, 96 rxn | 23 | ASXL1, CALR, CEBPA, CSF3R, DNMT3A, EZH2, FLT3, HRAS, IDH1, IDH2, JAK2, JAK3, KDM6A, KIT, MPL, NPM1, RUNX1, SETBP1, SF3B1, SRSF2, TET2, TP53, U2AF1 |
Bold indicates whole CDS coverage
IDT is committed to providing quality products to researchers working on the cutting edge of scientific discovery. The xGen Monkeypox Virus Amplicon Panel was designed as part of the next generation sequencing (NGS) Tech Access program at IDT, which is intended to accelerate innovation by enabling earlier access to our most advanced research tools. Tech Access products have not been through our standard, rigorous development cycle. These products are particularly well suited for researchers who require the most up-to-date technology to unlock new discoveries.
The current variants of monkeypox that are circulating have a genome of nearly 200 kb double-stranded DNA [1]. Surveillance of the virus and any potential mutations have gained international support due to the lessons learned through the COVID-19 pandemic. The IDT xGen Monkeypox Virus Amplicon Panel helps to enable researchers to track monkeypox strains, including potential new variants, by next generation sequencing.
DNA-to-sequencer in 2.5 hours
The workflow for the xGen Monkeypox Virus Amplicon Panel starts with extracted viral DNA (Figure 1). You can then generate an NGS library in a single tube using tiled primer pairs designed to target 184 kb of the monkeypox genome. Primers were designed for the currently circulating strain of the monkeypox virus (NCBI accession number ON568298 [1]), which allows you to generate overlapping amplicons in a single-tube, PCR 1 + PCR 2 workflow. If pooling multiple samples for NGS, the xGen Amplicon Core Kit includes the reagents for Normalase™ technology, the proprietary enzymatic normalization step that reduces hands-on time needed for manual normalization. Specifications of the Tech Access, xGen Monkeypox Virus Amplicon Panel are found in Table 1.
Table 1. Features of the xGen Monkeypox Virus Panel.
| Features | Specifications |
|---|---|
| Design coverage and panel information |
|
| Input Material |
|
| Time | ~2.5 hours for viral DNA-to-library |
| Multiplexing capability | Up to 1536 UDIs |
| Compatible with other indexes? | Yes |
| Recommended depth |
Strain identification or variant calling: 500K reads per library |
The xGen 16S Amplicon Panel v2 and the xGen ITS1 Amplicon Panel offer a reliable NGS workflow that provides coverage and NGS quality data on Illumina® sequencing platforms. These kits leverage multiplex PCR technology, enabling library construction from DNA using tiled primer pairs to target V1-V9 variable rRNA regions and the ITS1 region, each with a single pool of multiplexed primer pairs.
These xGen Amplicon Panel kits utilize multiple overlapping amplicons in a single tube, using a rapid, 2-hour sequencing workflow to prepare ready-to-sequence libraries for research studies (Figure 1). The single pool multiplex PCR workflow generates robust libraries, even from low-input quantities. The libraries may be quantified with conventional methods such as Qubit® (Thermo Fisher Scientific) or Bioanalyzer® (Agilent) and normalized by manual pooling or may be normalized enzymatically with the included xGen Normalase™ reagents. (Note: using the Normalase reagent adds additional time to complete the workflow).
In addition, both the xGen 16S Amplicon Panel v2 and the xGen ITS1 Amplicon Panel facilitate NGS analysis of complex microbial communities (e.g., bacteria, archaea, fungi) using single primer pools that target the 16S rRNA gene (variable regions 1–9) and ITS1 region, respectively. The panels can be customized with additional targets, including antibiotic resistance or virulence genes, allowing sub-genera level identification and functional analysis.
Figure 1. xGen Amplicon Panels have a single tube workflow that completes in 2 hours. Creating an NGS library starts with multiplex PCR. Your panel is combined with the DNA sample to amplify the targets of interest. The samples are then amplified with indexing primers to create a functional dual indexed library. As an optional step, the xGen Normalase reagent can be used after pooling multiple libraries to ensure each is equally represented in the final sample for the flowcell.
The xGen Sample ID Amplicon Panel is a streamlined amplicon NGS prep kit that includes 95 primer pairs targeting exonic single nucleotide polymorphisms (SNPs) with high minor allele frequency (MAF) and 9 amplicons to determine sex. This panel is compatible with circulating, cell-free DNA (cfDNA), as well as formalin-fixed paraffin embedded (FFPE) samples. With the advent of liquid biopsy assays of circulating nucleic acids in oncology research studies, proper tracking of samples has become increasingly critical. This panel represents a powerful and reliable method for tracking and identifying markers in a variety of sample types including liquid biopsy samples.
The power of discrimination of this panel is over 1 in 85,000 [1], making this product suitable for longitudinal studies and scenarios in which genetic fingerprinting is relevant to research design and analysis in various areas including genotyping, research on noninvasive prenatal testing (NIPT), research on transplant rejection, and others. This product is a complete kit that includes all elements necessary to generate multiplex libraries compatible with Illumina® sequencing platforms, including primer pairs and indexed sequencing adapters.
As seen in Figure 1, xGen Sample ID Amplicon Panels have a single tube workflow that is done in as little as 2 hours.
Figure 1. xGen Sample ID Amplicon Panels have a single tube workflow that is done in as little as 2 hours. Creating an NGS library starts with multiplex PCR. Your panel is combined with the DNA sample to amplify the targets of interest. The samples are then amplified with indexing primers to create a functional dual indexed library. As an optional step, the xGen Normalase™ reagent can be used after pooling multiple libraries to ensure each is equally represented in the final sample for the flowcell.
Specifications
| Features | xGenTM Sample ID Amplicon Panel |
|---|---|
| Input DNA required | 10–25 ng |
| Time required | 2 hours |
| Number of amplicons | 104 |
| Amplicon size | 120–160 (Average 145 bp) |
| On target percentage | >95% |
| Coverage uniformity at >20% of mean | >95% |
| Multiplexing on MiSeq® v2 Nano at 500X depth | 38 Samples |
| Multiplexing on MiSeq® v2 at 750X depth | 384 Samples |
| Sample compatibility | cfDNA and FFPE |
Using next generation sequencing (NGS) to analyze SARS-CoV-2 can help researchers understand COVID-19. IDT, in partnership with the ARTIC Network, is offering their latest design of the ARTIC SARS-Co-V-2 Amplicon Panel to support global availability of this important research tool.
The ARTIC network—a group of researchers who are developing a system for processing different virus samples to generate public health information—has developed primer sequences, experimental recommendations, and bioinformatic resources to facilitate NGS analysis of the SARS-CoV-2 genome. Their primer design pipeline (PrimalSeq), previously deployed for other infectious disease sequencing, was especially useful early in the SARS-COV2 pandemic [1].
ARTIC provides a detailed experimental protocol featuring a set of primers for amplicon sequencing to quickly sequence SARS-CoV-2 [2]. They also maintain a resource that alerts researchers to updates in the design [3]. An adaptative protocol that allows for sequencing on Illumina sequencers have been developed by others [4].
The sequence of the SARS-CoV-2 genome can enable research in many areas, including:
- Origin identification
- Mutations and viral evolution rates
- Transmission routes of variants
- Population surveillance, such as wastewater-based epidemiology.
University of Birmingham
ARTICnetwork
Workflow
The ARTIC SARS-CoV-2 Amplicon Panel workflow includes cDNA synthesis, amplicon generation, and cleanup. The final amplicons average 400 bp require 2 x 250 bp read lengths on either the NovaSeq™ or MiSeq™ sequencers (Illumina).
Table 1. Specifications for the ARTIC SARS-CoV-2 Amplicon Panel
| Recommended Ct value | Coverage | Contiguous amplicons | Coordinates covered |
|---|---|---|---|
| <30 | 99.7% | 99 | 50-29,827 |
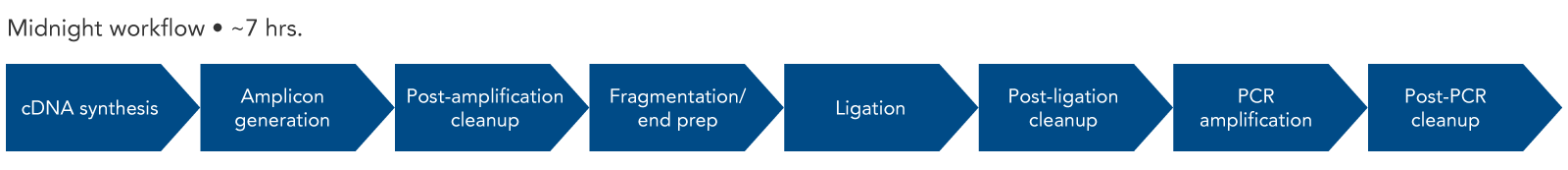
The Midnight Panel was created by Drs. Nikki Freed and Olin Silander of Massey University in New Zealand [1] and recently updated by Dr. John Tyson and Tracy Lee of the British Columbia Centre for Disease Control. This panel consists of 29 amplicons of approximately 1200 bp in length (Figure 1). The reduced number of amplicons allows for more consistent sequencing across the genome than would be achievable with a higher number of amplicons.
Figure 1. SARS-CoV-2 Midnight Amplicon Panel amplicons. The multiplex primer panel consists of two pools. The spacing is designed to tile across the entire SARS-CoV-2 genome, generating 29 amplicons approximately 1200 bp in length.
Since the primers only anneal to a total of 4.5% of the viral genome, the chance that viral mutations will disrupt primer annealing is lower. This results in a lowered chance of amplicon dropouts.
The Ct value for the amplicon sequencing reaction is an important metric since the higher the Ct, the lower the number of viral genomes in the sample. Samples with less viral genomic material pose difficulty for primer annealing and amplification.
In our experiment, contrived samples were prepared by mixing inactivated SARS-CoV-2 viral material into a negative background matrix of a negative nasopharyngeal (NP) swab in viral transport medium (VTM), with two samples per Ct value. Results show that when the SARS-CoV-2 Midnight Amplicon Panel and xGen DNA Library Prep Kit EZ are combined, the recommended Ct value is <32 which="" has="" been="" shown="" to="" sequence="" 97="" of="" the="" sars-cov-2="" genome="" with="" sufficient="" depth="" identify="" variants="" table="" 1="" p="">
Table 1. SARS-CoV-2 Midnight Amplicon Panel and xGen DNA Library Prep Kit EZ metrics.
| Recommended Ct value | Coverage (%) | Contiguous amplicons (count) | Coordinates covered |
|---|---|---|---|
| <32 | 97% | 29 | 30–28,985 |
Workflow
Since the length of the amplicons created by the SARS-CoV-2 Midnight 1200 Amplicon Panel are 1200 bp, Illumina® sequencing platforms are not compatible with this workflow. To use Illumina sequencing platforms, the SARS-CoV-2 amplicons generated with the Midnight panel can be fragmented using the xGen DNA Library Prep Kit EZ (with enzymatic DNA fragmentation). An overview of this workflow shows the steps necessary for library construction (Figure 2). IDT offers many of the reagents for the workflow, from the SARS-CoV-2 Midnight Panel and the components for library construction, to Nuclease-Free Water.
Figure 2. Expected experimental workflow for SARS-CoV-2 genome sequencing using the SARS-CoV-2 Midnight Amplicon Panel in combination with the xGen DNA Library Prep Kit EZ. By using the xGen DNA Library Prep Kit after the SARS-CoV-2 Midnight Amplicon Panel, the original 1200 bp fragments are converted into a library that is compatible with Illumina sequencers and chemistry.
For labs with access to Oxford Nanopore Sequencing instruments, the amplicons created using the SARS-CoV-2 Midnight Amplicon Panel can be prepared using one of the barcoding kits available directly from Oxford Nanopore (e.g., Rapid Barcoding Kit, SQK-RBK004).
The IDT xGen HS EGFR Pathway Amplicon Panel covers contiguous regions of the EGFR gene, and hotspot coverage of BRAF, KRAS, and NRAS with a panel of amplicon-generating primers. The final amplicons average only 136 bp making them compatible with DNA fragments from cell-free DNA (cfDNA) and/or FFPE DNA. The final target size of this panel is 1.5 kbp.
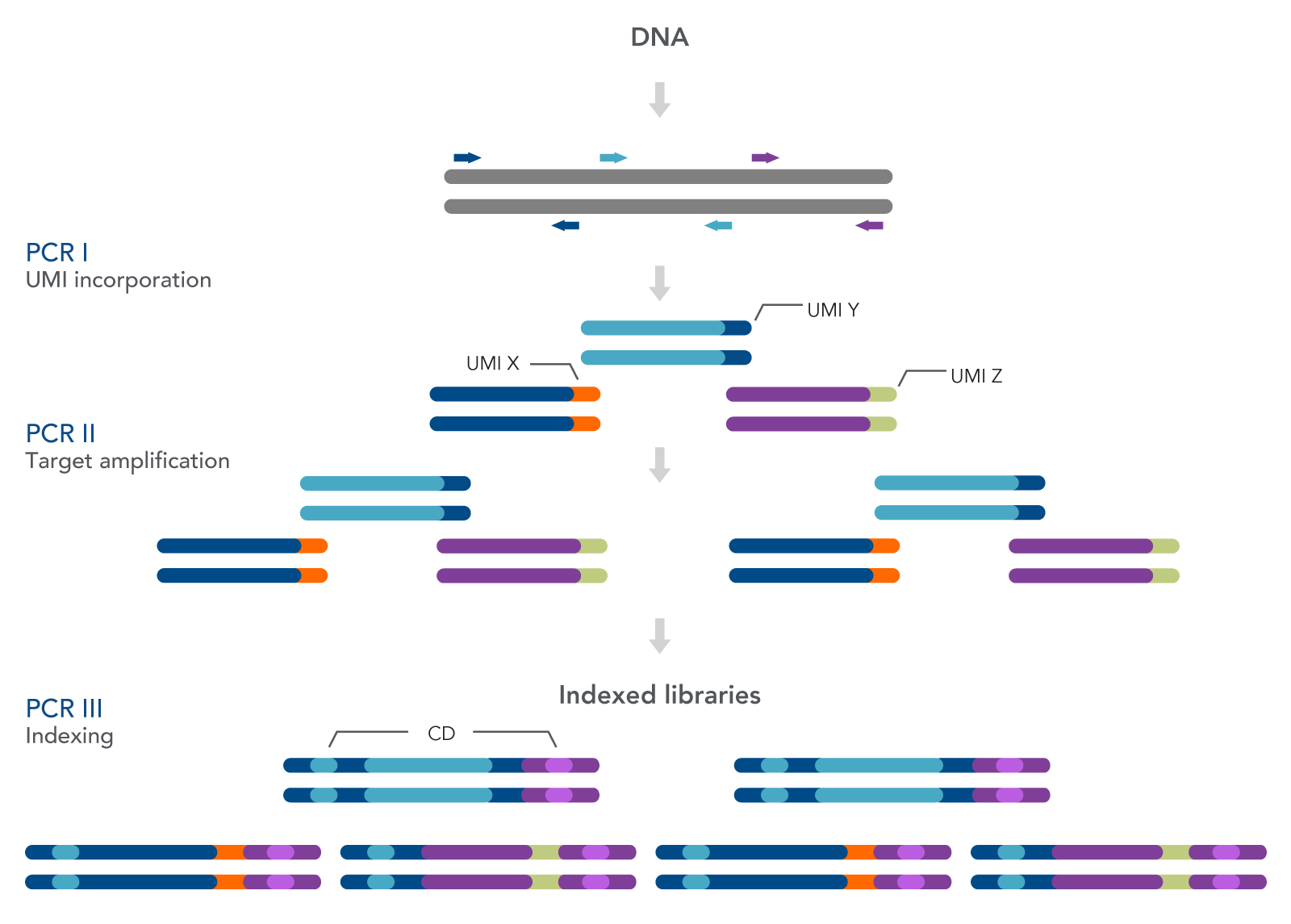
As seen in Figure 1, this workflow includes three PCR steps—one for the incorporation of unique molecular identifiers (UMIs) (PCR I), one for target amplification (PCR II), and one for the addition of combinatorial dual indexed adapters (PCR III), enabling multiplexing of up to 96 unique libraries. Bead-based SPRI® cleanups (Beckman Coulter) are used to purify the sample by removing unused oligonucleotides and changing buffer composition between steps.
Figure 1. The xGen HS EGFR Pathway Amplicon Panels are used to prepare indexed Illumina®-compatible libraries from cfDNA or FFPE DNA. The workflow for preparing the final targeting sequencing libraries includes PCR I, PCR II, and PCR III. The first amplification uses the panel of primers to amplify the targeted genes from the EGFR pathway. The primers also incorporate UMIs. A second amplification step increases the total number of fragments for indexing, while the third PCR completes the indexing.
PRODUCT DATA
To evaluate the xGen HS EGFR Pathway Amplicon Panel, two DNA standards from Seracare were used. These standards contain mutations with known allele frequencies at either 0.5% or 0.25%. Duplicate SeraSeq® ctDNA Reference Material AF (LGC SeraCare) samples at 10 ng were generated with the 0.5% standard and duplicate SeraSeq® ctDNA Reference Material AF (LGC SeraCare) samples at 20 ng were generated with the 0.25% standard, a total of 4 libraries are shown. As seen in Table 1, the observed allele frequency (AF) aligned with the expected allele frequency for the mutations in the reference cfDNA that were targeted by the xGen HS EGFR Pathway Amplicon Panel. Samples were sequenced on a MiniSeqTM (Illumina).
Table 1: Observed vs. expected allele frequency in NGS libraries prepared using the xGen HS EGFR Pathway Amplicon Panel
| 10 ng SeraSeq cfDNA with AF of 0.5% | 20 ng SeraSeq cfDNA with AF of 0.25% | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Mutation | Chr | Position | Expected AF | Observed AF (Rep 1) | Observed AF (Rep 2) | Expected AF | Observed AF (Rep 1) | Observed AF (Rep 2) |
| BRAF | p.V600E | 7 | 140453136 | 0.5% | 0.19% | 0.45% | 0.25% | 0.18% | 0.18% |
| EGFR | p.T790M | 7 | 55249071 | 0.5% | 0.43% | 0.57% | 0.25% | 0.33% | 0.36% |
| EGFR | p.L858R | 7 | 55259515 | 0.5% | 0.86% | 0.43% | 0.25% | 0.26% | 0.25% |
| KRAS | p.G12D | 12 | 25398284 | 0.5% | 0.59% | 0.47% | 0.25% | 0.20% | 0.48% |
| NRAS | p.Q61R | 1 | 115256529 | 0.5% | 0.91% | 0.56% | 0.25% | 0.30% | ND |
| EGFR | p.E746-A750 (delELREA) | 7 | 55242265 | 0.5% | 0.24% | 0.61% | 0.25% | 0.11% | 0.07% |
| EGFR | p.D770-N771 (ins) | 7 | 55249012 | 0.5% | 0.43% | 0.57% | 0.25% | 0.32% | 0.36% |
xGen™ Amplicon Core Kit
Targeted amplicon library prep kit for contiguous coverage (overlapping amplicons), comprises multiplexed PCR, indexing PCR, and Normalase™ reagents (target-specific panel and indexing primers sold separately).
xGen™ COVID Amplicon Panels
COVID panels that utilize multiple overlapping amplicons in a single tube, comprises a premixed target-specific primer pool.
The xGen SARS-CoV-2 Amplicon Panels enable identification of SARS-CoV-2 variants from research samples such as nasopharyngeal/oropharyngeal swabs, sputa, stool, and wastewater. The xGen ACE2 Gene Amplicon Panel amplifies the angiotensin converting enzyme 2 gene which has been proven to be the SARS-CoV-2 spike protein receptor [1]. The xGen SARS-CoV-2 Sgene panel amplifies the viral gene for the spike protein only.
The xGen SARS-CoV-2 Amplicon Panels support:
- 99.7% genomic coverage†
- Full genomic coverage of lineages from a single primer set
- Sequence data from viral titers as low as 10–100 viral copies
- cDNA-to-sequencer in 3 hours
- Up to 1536 UDIs
xGen™ Amplicon UDI Primers
Premixed Normalase unique dual index primer pairs for sample indexing during library amplification and conditioning for Normalase enzymology, 1536 ten base index pairs dispensed into 16 96-well plates, Reagent R7 NOT provided, instead uses reagent supplied in the xGen Amplicon Core Kit.
xGen™ PEG NaCl Buffer
Buffer used during ''with bead'' purification steps when DNA is eluted into the next reaction mixture and the beads are reused. Supplied with the xGen Amplicon Core Kit but also sold separately if additional buffer is needed.
Gen™ Amplicon CDI Primers
Individual combinatorial dual index primers for sample indexing during library amplification, 20 eight-base index primers dispensed separately for 96-plex combinations.
xGen™ Oncology/Inherited Disease Amplicon Panels
Oncology and infectious disease panels that utilize multiple overlapping amplicons in a single tube; comprises a premixed target-specific primer pool.
- Compatible with Illumina® sequencing platforms
- Designed for research on germline and somatic variant identification, or targeted sequencing
- Offers overlapping amplicons in a fast, easy, single-tube workflow
xGen™ Monkeypox Virus Amplicon Panel
Monkeypox Virus panel that utilizes multiple overlapping amplicons in a single tube, comprises a premixed target-specific primer pool.
Monkeypox has been termed a global health emergency. For researchers interested in tracking the evolution of the monkeypox virus genome, it can be difficult to get sufficient coverage. The xGen Monkeypox Virus Amplicon Panel provides a single-tube, two-step PCR amplification workflow with primer sets designed to create amplicons across the entire genome.
The xGen Monkeypox Virus Amplicon Panel supports:
- Comprehensive coverage from positions 6760–190,905 (ITRs not included)
- Sequence data from viral titers as low as 300 viral genome copies
- Single-tube workflow
- Viral DNA-to-sequencer in ~2.5 hours
- Up to 1536 UDIs
- Super amplicon technology
xGen™ Metagenomics Amplicon Panels
Metagenomics panels that utilize multiple overlapping amplicons in a single tube; comprises a premixed target-specific primer pool.
- The xGen 16S rRNA v2 Amplicon Panel targets V1–V9 variable regions of the 16S rRNA gene
- The xGen ITS1 Amplicon Panel targets the internal transcribed spacer regions of the rRNA genes
- Integrated library normalization enables streamlined library balancing and pooling without the need to quantify samples
- Unique amplicon chemistry generates diverse clusters without PhiX or phased primers, leading to more efficient sequencing (Figure 4)
- Compatible with Illumina® sequencers and read lengths
- Simple, single tube, 2-hour workflow
xGen™ Sample ID Amplicon Panel
Sample ID panel that utilizes multiple overlapping amplicons in a single tube; comprises a premixed target-specific primer pool.
- Easily track samples within and between studies
- Ideal for confirming research on tumor/normal pairs
- Power of discrimination over 1 in 85,000 [1]
- Reliable calling of germline variants
- Complements both WGS and exome sequencing for sample tracking
- On-target coverage uniformity >95%
- Leverages the high fidelity of Illumina® platforms
- Complete library generation in a single kit
- Reduce sequencing cost with 1536 index pairs
ARTIC nCoV-2019 Amplicon Panel
Predesigned panel designed in collaboration with the ARTIC network targeting the SARS-CoV-2 genome.
SARS-CoV-2-Midnight-1200 Amplicon Panel
The updated Midnight Panel V2 was designed by Dr. John Tyson and Tracy Lee (BC Centre for Disease Control) together with Dr. Nikki Freed (Massey University and University of Auckland) and Dr. Olin Silander (Massey University)
- Amplifies the SARS-CoV-2 genome for sequencing using premixed multiplex PCR primers
- Provides data for understanding SARS-CoV-2 viral evolution and the epidemiology of COVID-19
- Data identifies SARS-CoV-2 variants, including alpha, beta, gamma, delta, mu, and omicron
- Panel generates 29 tiled amplicons, approximately 1200 bp in length
- Panel is optimized for high throughput applications or automated workflows
- Samples with Ct values <32 are="" recommended="" for="" complete="" genome="" sequencing="" coverage="" based="" on="" initial="" testing="" li="">
xGen™ HS Amplicon Panel
EGFR amplicon panel utilizing UMIs for identification of mutations occurring at 0.25% allele frequency. Custom panels available upon request.
- Identifies SNVs and indels down to 0.25% allele frequency
- Compatible with cfDNA and FFPE DNA
- Amplifies from 10–50 ng of cfDNA
- Makes Illumina®-compatible libraries within 3 hours
- Supports research in oncology, graft vs. host disease, and single nucleotide variants in the EGFR pathway
 Forgalmazott termékeink gyártói - keressen gyártó szerint a logóra kattintva
Forgalmazott termékeink gyártói - keressen gyártó szerint a logóra kattintva